Enzymes are vital biological catalysts for a multitude of processes, from cellular metabolism to industrial manufacturing. The applications of artificial…
Enzymes are vital biological catalysts for a multitude of processes, from cellular metabolism to industrial manufacturing. The applications of artificial intelligence for enzyme generation is an exciting field of research with direct applications in the life sciences. Advances in these scientific challenges are a critical necessity to further advance drug discovery, environmental science, and bioengineering.
Currently, only a tiny fraction of Earth’s vast array of life forms has been sequenced, hindering the broader application and generalization of machine learning algorithms within the complex realm of sequence design. Improved methods for functional annotation are a vital component in enzyme research, enabling the identification and characterization of the functions of newly discovered enzymes. This is key to understanding complex biological processes and enhancing the data used for generative workflows.
Basecamp Research, a London-based Bio-AI company and NVIDIA Inception member, recently used NVIDIA GPUs to train a Hierarchically Fine-tuned Nearest Neighbor method (HiFi-NN). This approach has shown significant improvements over existing models in recall, precision, and F1 scores, surpassing the state-of-the-art (SoTA) in enzyme annotation by over 15%.
Unique data collected in global expeditions
Basecamp Research tackles the most complex biological design challenges across the biotech industry. Existing datasets have significant shortcomings:
- Small representativity, covering only 0.001% of life on earth
- No consistent metadata
- Lack of stakeholder consent and engagement before data collection
Basecamp opted to develop its proprietary biological data resource through biodiversity partnerships with nature parks across five continents and 23 countries. They sent their scientists on worldwide expeditions to discover new genomes, enzymes, and biological relationships from the most extreme and extraordinary biomes.
In under two years, they created BaseGraph, the largest knowledge graph of natural biodiversity, containing over 5.5B relationships with a genomic context exceeding 70 kilobases per protein. Their extensive long-read sequencing is complemented by comprehensive metadata collection, enabling them to link proteins of interest to specific reactions and desired process conditions.
Basecamp Research’s AI strategy is data-centric for two reasons: their proprietary data enhances model performance in a rapidly commoditizing algorithmic landscape and significantly compensates for the lack of diversity in publicly available data.
Their knowledge graph, built from the ground up, captures and re-creates the complexities of four billion years of protein evolution in nature. This data advantage enables their AI and product teams to outperform SoTA annotation and design models, addressing complex design challenges in biotech industries, from gene-writing therapeutics to plastic degradation.
In silico functional annotation
To tackle the challenge of in silico functional annotation of proteins and enzymes, Basecamp Research’s deep learning (DL) team developed HiFi-NN search. HiFi-NN annotates protein sequences with enzyme commission (EC) numbers beating the current bioinformatics tool of choice (blastp), as well as other SoTA DL models, including CLEAN, in precision and recall (Table 1).
MethodRecallPrecisionF1-scoreECPred0.01970.01970.0197DEEPpre0.04030.04150.0386DeepEC0.07240.11840.0846ProteInfer0.13820.24340.1662ProteinVec0.29610.49010.3378BLASTp0.37500.50830.3852DeepECtransformer0.30260.52630.3511CLEAN0.46710.58440.4947HiFi-NN (Swissprot)0.57240.55050.5304HiFi-NN (Swissprot + 3M curated sequences)0.59210.66570.6015Table 1. HiFi-NN results compared to existing tools and other SoTA DL models
BCR 3M refers to the version of the model retrained with 3M selected, environmentally diverse sequences from Basecamp Research’s BaseGraph.
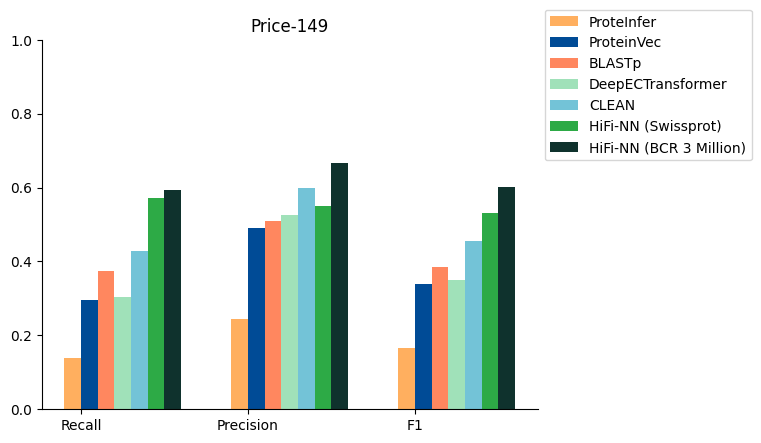 Figure 1. HiFi-NN results compared to SoTA models in Enzyme Functional Annotation on the price-149 enzyme benchmarking dataset
Figure 1. HiFi-NN results compared to SoTA models in Enzyme Functional Annotation on the price-149 enzyme benchmarking dataset
Developed in collaboration with advisors Noelia Ferruz and Kevin Yang, HiFi-NN was accepted by NeurIPS, a prestigious AI conference, for presentation at their Machine Learning for Structural Biology workshop in December 2023.
The model uses contrastive learning of EC numbers and the inherent hierarchy of the enzyme commission annotation system for natural augmentation. Trained on eight NVIDIA A100 GPUs on a Lambda Labs instance with CUDA version 11.8 and NCCL version 2.14.3, it employs PyTorch Lightning for distributed-data parallel training. Experiment management and tracking are done through the Hydra framework and Weights and Biases. The model boasts over 3M parameters.
HiFi-NN’s superior performance in SoTA annotation methods is attributed to using the hierarchical nature of enzyme function represented in the EC numbering system and supplementing the training set with proprietary sequences from Basecamp’s knowledge graph.
Proprietary sequences
The proprietary Basecamp sequences that HiFi-NN is supplemented with originate from environments spanning five continents, and a 110o C temperature range to ensure as much sequence and environmental diversity in the training set as possible. As a result, HiFi-NN outperforms all SoTA models on benchmarking datasets and performs particularly well on protein sequences from functional dark matter, that is, those sequences with low similarity to any known enzymes.
In fact, the Basecamp team used HiFi-NN to annotate a large representative portion of the MGnify microbial protein database that was previously not annotated.
In addition to outperforming all previous annotation models, HiFi-NN is also particularly easy to use and generates annotation labels at great speed. For example, it annotates the entire human proteome within 24 minutes on a single NVIDIA A100 GPU.
Fast enzyme identification with Johnson Matthey
Breakthroughs like HiFi-NN, which enhance our capacity to predict the physical properties of biological entities, are set to reduce the need for extensive screening of candidates using resource-heavy lab methods.
Basecamp Research’s partnership with an FTSE100 chemicals company, Johnson Matthey, underscores the importance of computational advancements in addressing industrial challenges. A notable project with this partner involved their researchers spending over a year and a half testing thousands of enzyme variants in their labs without success.
Johnson Matthey’s objective was to find an enzyme with broad specificity capable of processing multiple bulky substrates, a more complex task compared to working with smaller substrates. Within a week, Basecamp Research employed its entirely in silico techniques to identify an enzyme (Figure 2) meeting these criteria, positioning it for potential commercialization.
The research group’s leader expressed admiration for Basecamp Research’s capability to rapidly discover and develop an enzyme, a task that had eluded them for years. This success laid the groundwork for an expanded collaborative effort on various enzyme development initiatives.
 Figure 2. Basecamp Research’s proprietary protein
Figure 2. Basecamp Research’s proprietary protein
Bolstering AI development in life sciences
Functional annotation plays a pivotal role for researchers, especially in practical scenarios:
- In drug discovery, it aids in the creation of targeted treatments by elucidating enzyme interactions within the body.
- In the realm of industrial biotechnology, it enables the bespoke design of enzymes, tailored for specific industrial applications, promoting more environmentally friendly production methods.
- It also offers crucial insights into evolutionary biology by revealing the developmental trajectory of enzymes across different species.
In essence, functional annotation, driven by machine learning, transcends its role as a mere scientific instrument. it acts as a catalyst for innovation and exploration across diverse sectors, including healthcare and environmental science.
Basecamp Research, along with other life sciences entities, is integrating its workflow with NVIDIA BioNeMo, a generative AI platform geared towards drug discovery. This platform streamlines and expedites the training of models. With BioNeMo, organizations can tailor and deploy AI models for various purposes, including 3D protein structure prediction, de novo protein and small molecule generation, property prediction, and molecular docking.
If you’re interested in exploring BioNeMo, opportunities are available through the BioNeMo Framework beta release or the API early access program., For more information, see the NVIDIA BioNeMo product page.
Source:: NVIDIA
