The upcoming 4.1 release of NVIDIA Parabricks, a suite of accelerated genomic analysis applications, goes further than ever before in accelerating sequencing…
The upcoming 4.1 release of NVIDIA Parabricks, a suite of accelerated genomic analysis applications, goes further than ever before in accelerating sequencing alignment and increasing the accuracy of deep learning variant calling. The release includes a new workflow for PacBio long-read data, featuring an accelerated Minimap2 tool and Google’s DeepVariant for full GPU-enabled, end-to-end analysis of PacBio data.
NVIDIA Parabricks is free to use with an option for paid enterprise support. It contains a variety of optimized and AI-based industry-standard genomic tools delivering up to 80x acceleration over CPU-based tools and reducing compute costs by up to 50%. A 30x whole genome can now be analyzed in just 16 minutes compared to ~24 hours on CPU, translating to the analysis of up to 30,000 whole genomes a year on a single server.
A quick look at Parabricks v4.1 features
- A new DeepVariant Re-Training tool, to enable anyone to re-train or fine-tune DeepVariant for their own data, enabling more accurate variant calling (available now on NGC).
- An end-to-end (FastQ-to-VCF) accelerated workflow for PacBio, which will be made available on the Parabricks workflows on GitHub, Terra.Bio, and other cloud platforms.
- New accelerated Minimap2 tool for alignment of PacBio’s long reads.
- New accelerated DeepVariant variant caller for PacBio data with an 8-minute runtime for a 30x whole genome on 2xA100 GPUs.
- Further acceleration of the short-read germline pipeline for a 30x whole genome in 16 mins on a DGX A100 GPU [8xA100 GPUs] compared to 21 mins in v4.0 and ~24 hours on CPU-only.
- Compatibility with the new NVIDIA H100 GPU, which includes a powerful DPX instruction for boosting dynamic programming algorithms like Smith-Waterman for local sequence alignment.
Sign up for notification of the Parabricks 4.1 release, or try the prerelease DeepVariant re-training tool.
Figure 1. Parabricks v4.1 optimization of the PacBio model for DeepVariant
Supporting long-read analysis
Long-read sequencing, the capability of sequencing significantly longer fragments of DNA, has multiple inherent advantages over traditional short-read sequencing. Most prominently, the reads are more easily assembled into the full genome.
Lower levels of ambiguity and alignment error make long-read sequencing better for more challenging parts of the genome (for example, highly repetitive regions) or for assembling a genome de novo (without a provided reference).
This has resulted in a multitude of improvements for the sequencing community, including a greater understanding of structural variants (large insertions, deletions, inversions, duplications, and so on). Structural variants can be pathogenic for diseases, such as Lou Gehrig’s disease (ALS), Parkinson’s disease, and cardiac diseases.
It has also finally enabled the scientific community to fully complete the human reference genome end-to-end, known as the telomere-to-telomere (T2T) genome released in 2022.
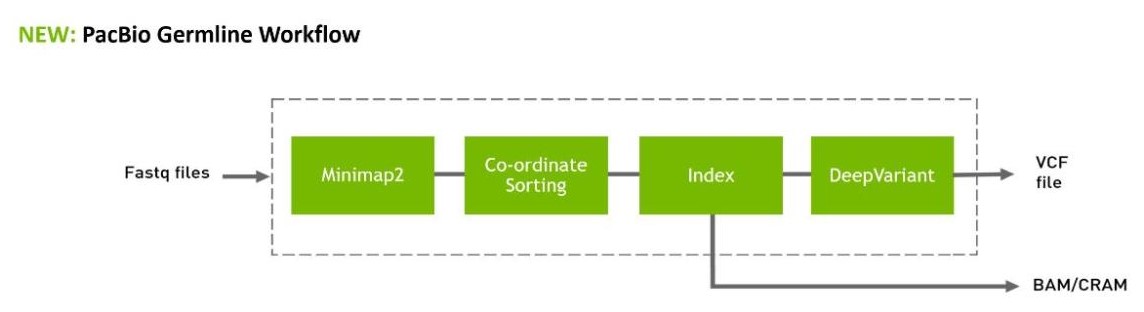 Figure 2. Long-read tooling and workflow available in Parabricks 4.1, with new Minimap2 and FastQ-to-VCF for PacBio
Figure 2. Long-read tooling and workflow available in Parabricks 4.1, with new Minimap2 and FastQ-to-VCF for PacBio
PacBio is a prominent leader in long-read sequencing. Their technology produces reads up to 25 kilobases in length (compared to short-read sequencing of
Source:: NVIDIA
