The quest for new, effective treatments for diseases that remain stubbornly resistant to current therapies is at the heart of drug discovery. This traditionally…
The quest for new, effective treatments for diseases that remain stubbornly resistant to current therapies is at the heart of drug discovery. This traditionally long and expensive process has been radically improved by AI techniques like deep learning, empowered by the rise of accelerated computing.
Receptor.AI, a London-based drug discovery company and NVIDIA Inception member, has successfully integrated NVIDIA BioNeMo cloud APIs with their end-to-end Computer Assisted Drug Discovery (CADD) platform. In doing so, the company has achieved significant performance improvements and cost savings by shifting major drug discovery tasks, such as virtual screening, ADMET prediction, and the ligand pose prediction, from traditional CPU-based processing to accelerated computing on the NVIDIA platform.
This post explores how Receptor.ai combined its Drug Discovery platform with NVIDIA BioNeMo, as well as the benefits of accelerated computing in ADMET prediction tasks.
NVIDIA BioNeMo as a platform enabler
NVIDIA BioNeMo cloud APIs pioneer the availability of advanced AI models key to drug discovery. Using foundation models for protein folding, molecular docking, and molecular generation, researchers, engineers, and drug hunters can scale and customize their development and deployment of large biomolecular generative AI applications through easy-to-use APIs. BioNeMo also includes a playground (Figure 1) serving as an experimental interface and a reference for developers to create their applications.
“NVIDIA’s BioNeMo provides a foundational layer of high-performance tools and state-of-the-art models, which can be easily customized and integrated into third-party drug discovery workflows,’ said Dr. Sergii Starosyla, CTO at Receptor.AI.
“Combining the power of BioNeMo with Receptor.AI’s drug discovery cloud platform allows us to get access to the latest deep learning techniques backed up by the power of the full stack of NVIDIA hardware and software accelerations. This opens up new opportunities for working with the most challenging drug targets,” Dr. Starosyla added.
Receptor.AI specializes in designing highly efficient therapeutics with an emphasis on selectivity and targeting protein-protein interactions (PPIs) using the power of an in-house hybrid intelligence drug discovery platform.
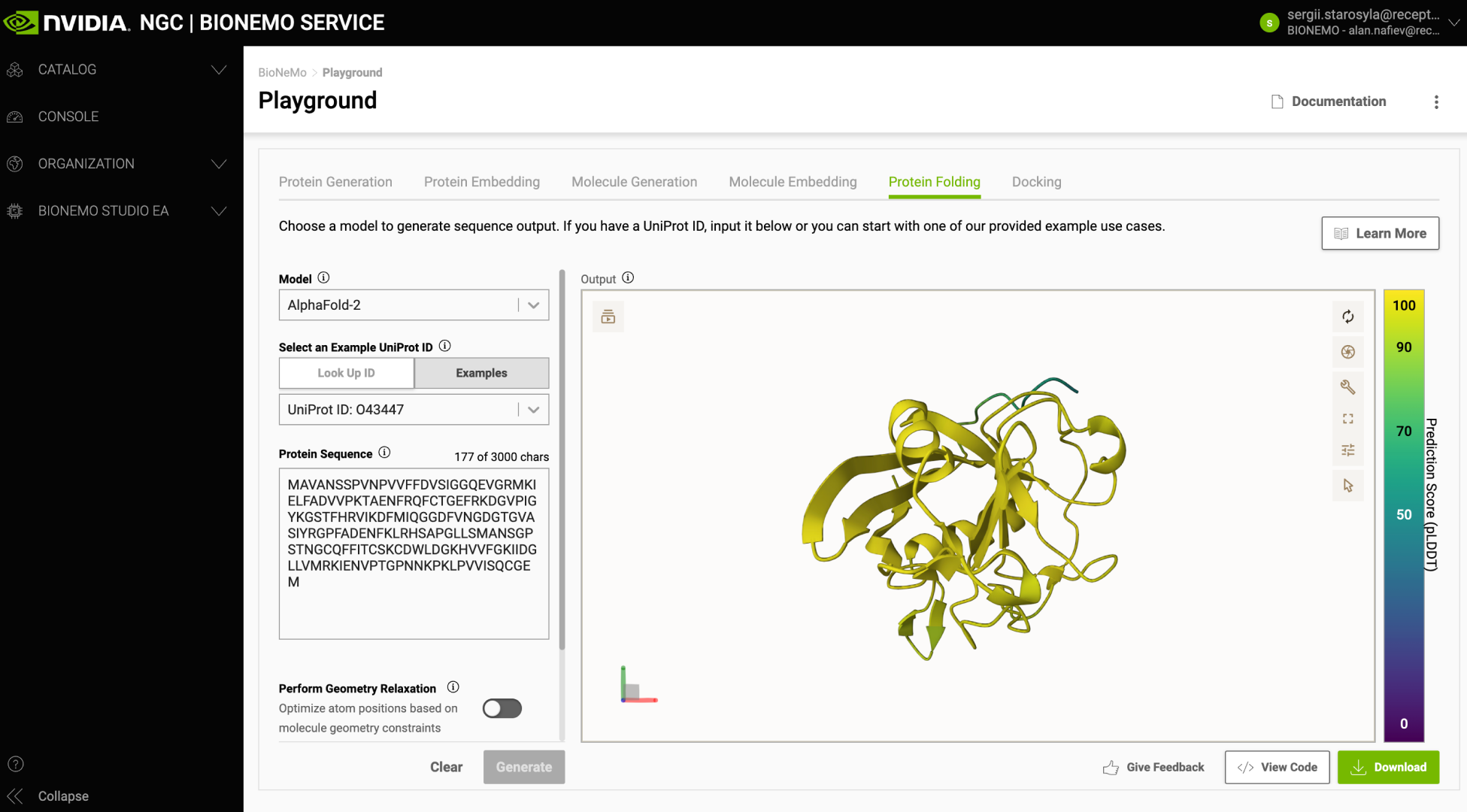 Figure 1. NVIDIA BioNeMo Playground for experimenting with models
Figure 1. NVIDIA BioNeMo Playground for experimenting with models
Diffusion-based models for biology and chemistry
Receptor.AI leverages diffusion-based generative AI to model drug-target interactions (DTIs), or how a protein in the body interacts with a drug. Through this system, they explore multibillion chemical spaces, factoring in the conformational flexibility of on-target and explicit off-target proteins, to ensure optimal identification of potential therapeutic compounds.
For example, Receptor.AI interaction models can predict whether a molecule interacts with a particular protein, indicating biological activity. Likewise, the selectivity prediction AI model assesses potential off-target interactions, and ranks its selectivity against thousands of human proteins. Receptor.AI’s platform also evaluates a compound’s absorption, distribution, metabolism, excretion, and toxicity (ADME-Tox) to ensure safety and efficacy, predicting up to 40 parameters.
Given the compute complexity involved in training and serving foundation models, accelerated computing platforms have become an essential component to operating them. One such model, the Evolutionary Scale Modeling (ESM) protein sequence encoding model available through the BioNeMo, provides input features for both the proprietary DTI and selectivity applications by Receptor.ai. For more information, see Language Models of Protein Sequences at the Scale of Evolution Enable Accurate Structure Prediction.
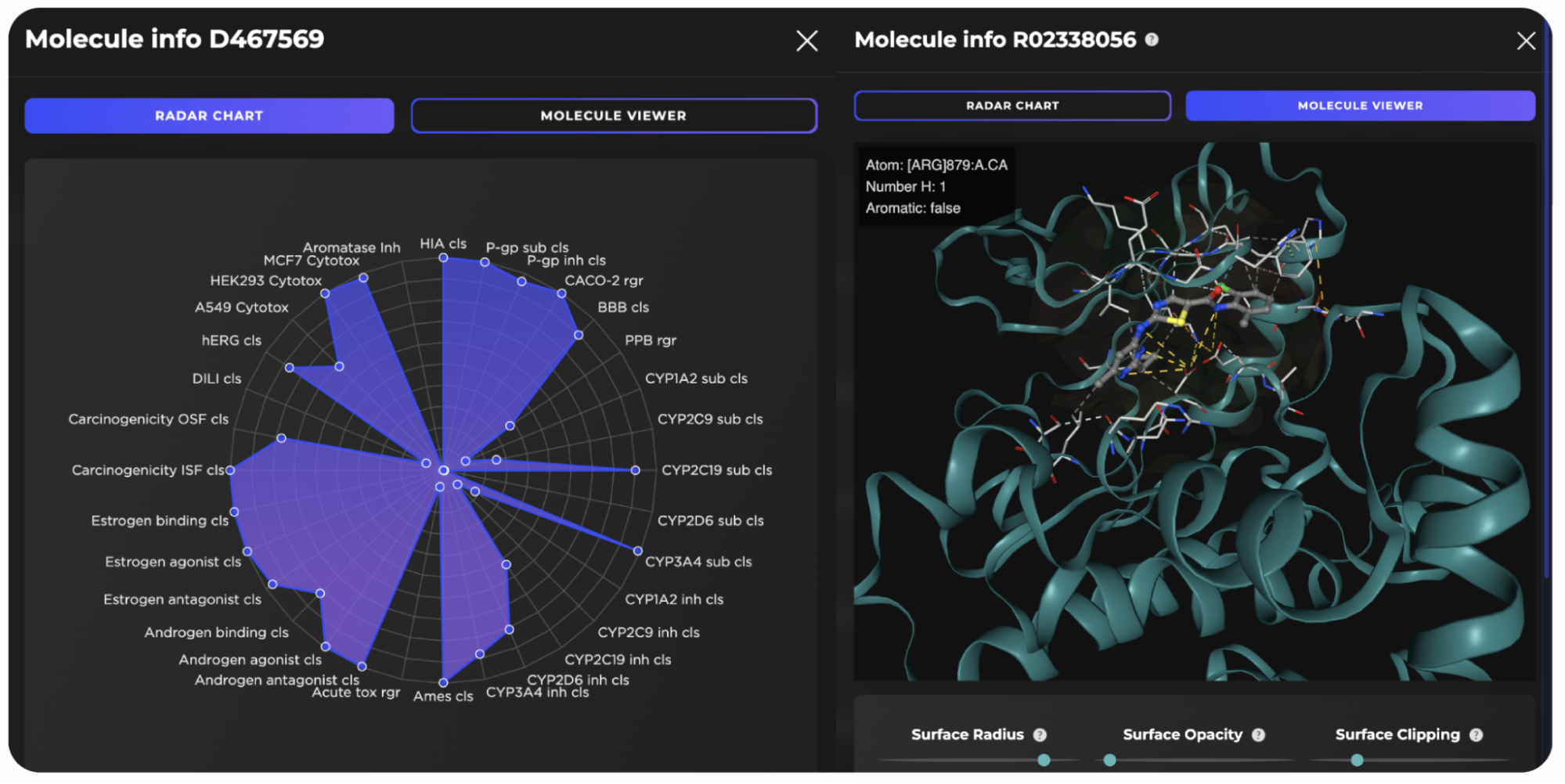 Figure 2. Web interface of the Receptor.AI drug discovery platform
Figure 2. Web interface of the Receptor.AI drug discovery platform
Drug design with foundation models
The Receptor.AI platform enables users to screen billions of compounds and assess the quality of the screening against a well-known target protein with high therapeutic potential. By combining their platform’s AI technologies with NVIDIA BioNeMo cloud APIs, Receptor.AI was able to streamline protein preparation, large-scale virtual screening, and docking techniques.
One Receptor.AI workflow is concerned with understanding long-chain polyunsaturated fatty acids (PUFA), pivotal for the proliferation of cancer cells. Given the importance of a human fatty acid desaturase 1 (FADS1, UniProt ID O60427) in PUFA biosynthesis and its involvement in cancer genesis and biology, this protein was identified as a promising drug target. For the selectivity assessment against FADS1, three homologous fatty acid desaturase domains were selected as potential off-targets: FADS2 (ID O95864), FADS3 (ID Q9Y5Q0), and FADS6 (ID Q8N9I5).
Receptor.AI has curated a vast library consisting of approximately 4.3 billion in silico-generated small molecule compounds for screening, including 25 known to bind FADS1 (known as ligands). This was used to assess the performance of Receptor.ai’s AI workflow on tasks to prioritize and rank compounds. Importantly, these ligands were excluded from the AI training set to maintain the integrity of their unbiased testing approach. Furthermore, Receptor.AI incorporated molecules computationally derived from known FADS1 ligands to demonstrate the robust generative AI capabilities of the BioNeMo and Receptor.AI platforms.
Preprocessed proteins and molecules for screening
Figure 3 outlines the scheme of the Virtual Screening experiment. The primary selection used the DTI and selectivity models, followed by the ADME-Tox assessment. DTI rank was used for proteome-wide selectivity assessment. BioNemo DiffDock AI docking was used to obtain the binding poses of shortlisted compounds. In the final stage, the Receptor.AI consensus function was used to rank the top candidate molecules.
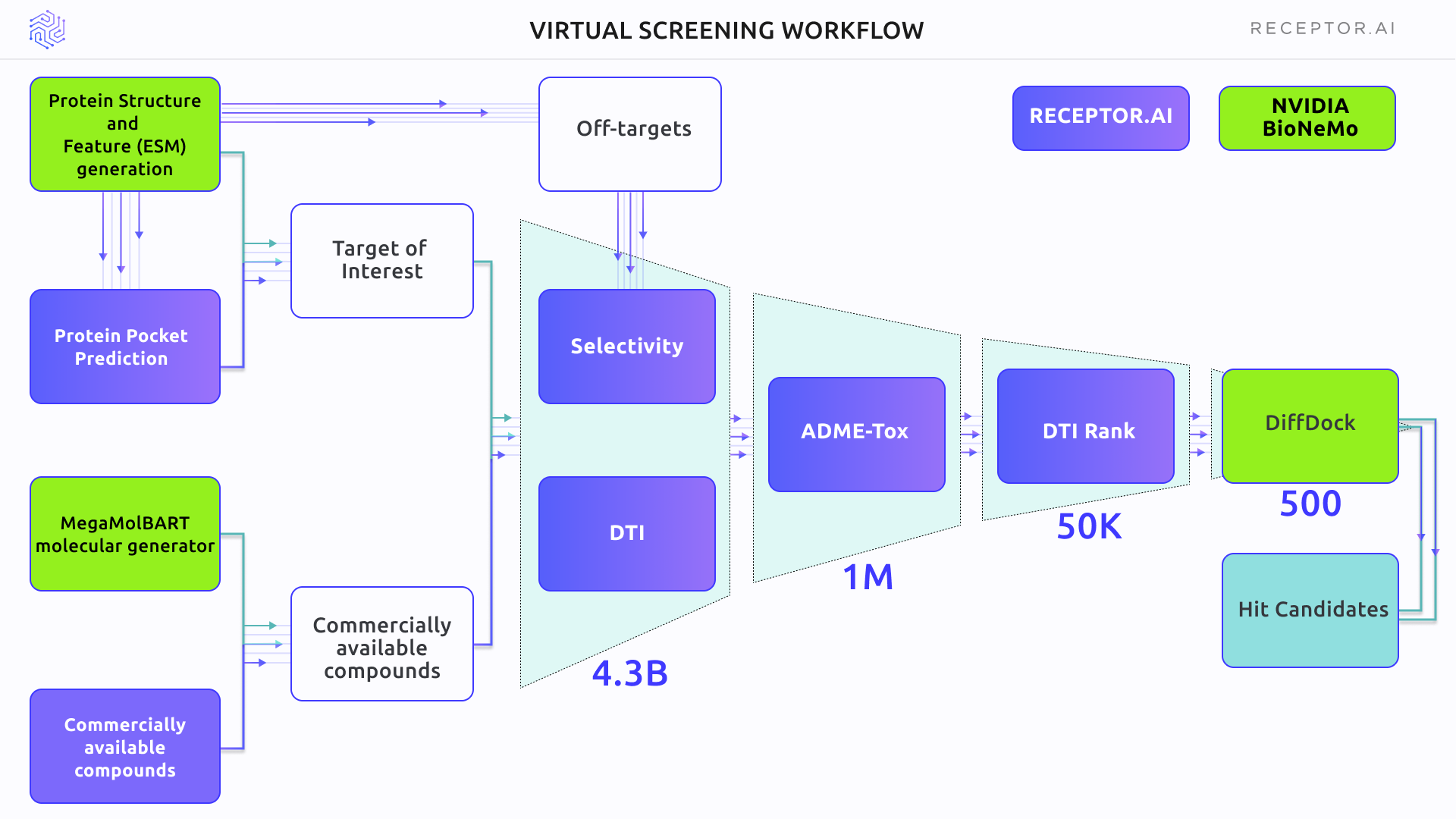 Figure 3. The general scheme of the experiment, including both BioNeMo (green blocks) and Receptor.AI (blue blocks) technology
Figure 3. The general scheme of the experiment, including both BioNeMo (green blocks) and Receptor.AI (blue blocks) technology
Protein preparation
The FADS1 protein does not have an experimental 3D structure available, fundamental for a drug discovery campaign. While its predicted structure is deposited in the AlphaFold Protein Structure Database, to demonstrate the BioNeMo AlphaFold module, it was recomputed in about 17 minutes with a high degree of confidence through BioNeMo cloud APIs (Figure 4). Such functionality is crucial when working with newly discovered or mutated proteins. For more details, see Highly Accurate Protein Structure Prediction with AlphaFold.
The ligand binding pocket was determined by the PUResNet AI model on a NVIDIA Tesla T4 GPU. Expectedly, the predicted pocket was located within the fatty acid desaturase domain in accordance with InterPro FADS1 annotation.
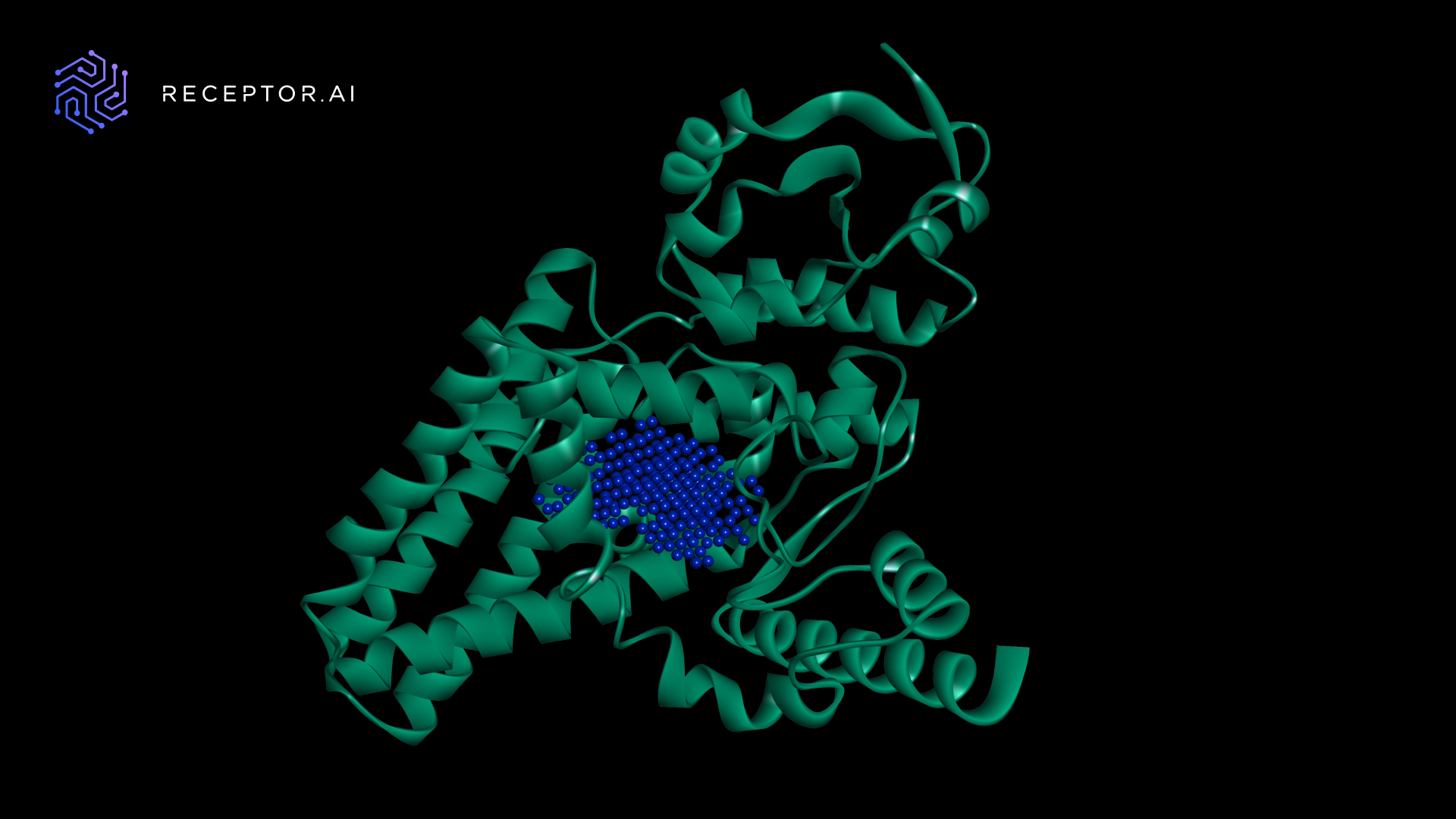 Figure 4. Predicted 3D structure of FADS1 and the binding pocket location (in blue)
Figure 4. Predicted 3D structure of FADS1 and the binding pocket location (in blue)
Using BioNeMo Cloud APIs, the ESM encodings of the target and three off-target proteins were computed. These required about 1.4 seconds to compute through the cloud APIs, compared to about 6.0 seconds on a CPU-only instance with 16 virtual CPUs.
Screening library preparation
Using the NVIDIA MegaMolBART model, 44 known compounds with experimentally determined activity for FADS1 (IC50 < 1K nM) from the ChEMBL database were used to generate 44K new molecules (1K per known compound). Figure 5 shows examples of generated molecules.
Receptor.AI mixed these 44K generated compounds into the 4.3 billion virtual library. The compounds from this library serve as tentative negative controls. They also added 25 randomly selected known ligands for FADS1 to the screening dataset to assess their ranking in the final results. This was done with the expectation that a considerable fraction of known ligands would occur on top of the final list of ranked hit candidates.
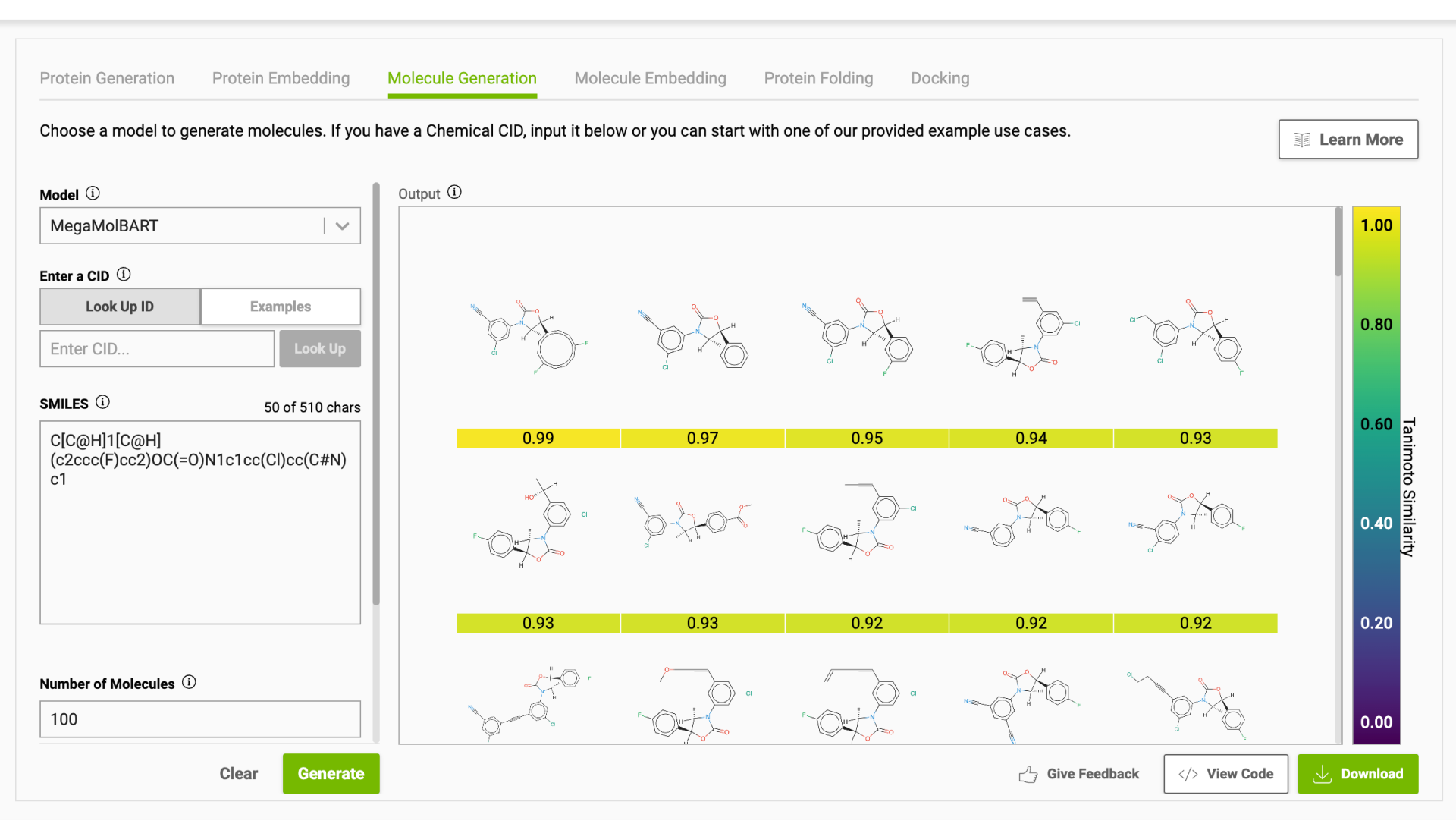 Figure 5. Examples of the molecules generated using MegaMolBART in the BioNeMo Playground
Figure 5. Examples of the molecules generated using MegaMolBART in the BioNeMo Playground
GPU-powered virtual screening
The Receptor.AI hit identification pipeline includes DTI, DTI rank, selectivity, and ADME-Tox modules (Figure 6). It was used to narrow the chemical space from about 4.3 billion compounds to the 500 most promising candidates. The screening process involves both CPU-intensive and GPU-intensive tasks. Table 1 compares the performance of a single NVIDIA T4 GPU against instances with 16 vCPUs for different stages of the virtual screening pipeline. Using GPUs consistently improves performance with speed-ups of 1.1x to 11.3x, depending on the task.
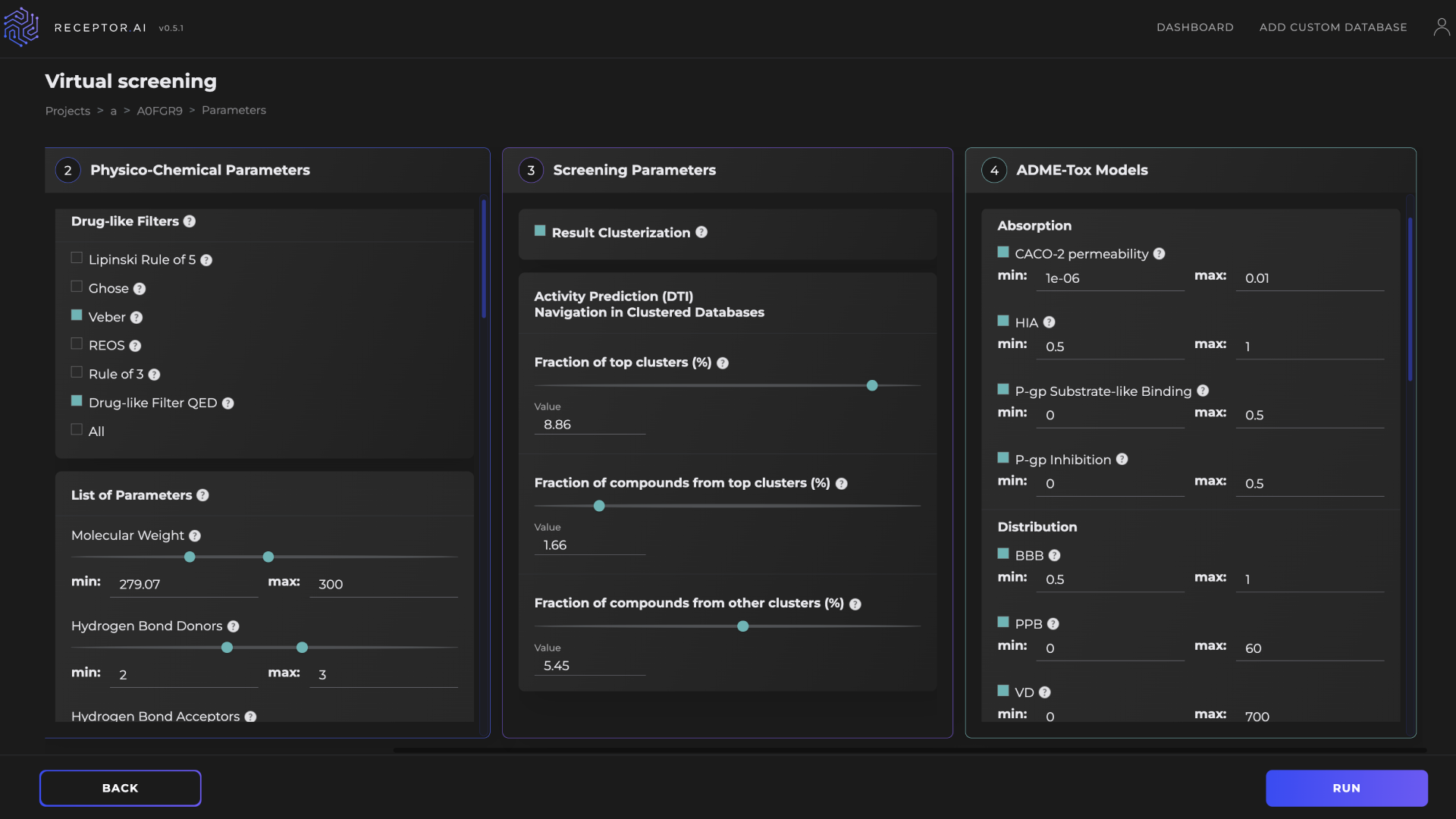 Figure 6. Receptor.AI platform virtual screening setup
Figure 6. Receptor.AI platform virtual screening setup
TechniqueMolecule #CPU run time (h)GPU run time (h)Acceleration factorDTI4.3 billion33.911.33Selectivity10 million10.91.1ADME-Tox1 million1.050.42.6DTI rank (against 9.3K proteins)50K6.10.87.6Total:42.0513.43Table 1. Inference times for the virtual screening library
The top 500 compounds identified by the Receptor.AI platform were then docked blindly to the FADS1 without a priori information about the location of the binding pocket. This process used the BioNeMo cloud API for DiffDock.
During protein preparation, the FADS1 protein structure has been confidently predicted in under 17 minutes using the BioNeMo AlphaFold module. The PUResNet AI model determined the ligand binding pocket.
During the screening library preparation, the ESM encodings of the target and three off-targets were computed faster through the BioNeMo Cloud API, which is backed by the same technology available through NVIDIA DGX Cloud, compared to virtual CPUs.
Lastly, the Receptor.AI hit identification pipeline, which involves both CPU-intensive and GPU-intensive tasks, used NVIDIA T4 GPUs. Specifically, the inference time for the 4.3 billion compound virtual screening library on the GPU was considerably reduced compared to instances with 16 vCPUs. This technological edge provided by the NVIDIA GPU significantly speeds up the processing time and improves the efficiency of the overall experiment.
Results
Receptor.AI has successfully identified several hit compounds from the known FADS1 ligands, as well as novel compounds with higher predicted affinity to FADS1, compared to the known ligands. Out of the 25 known FADS1 ligands, four were detected in the top 10 hit candidates. Almost half of all known ligands (12 of 25) were found in the top 1K (Figure 7).
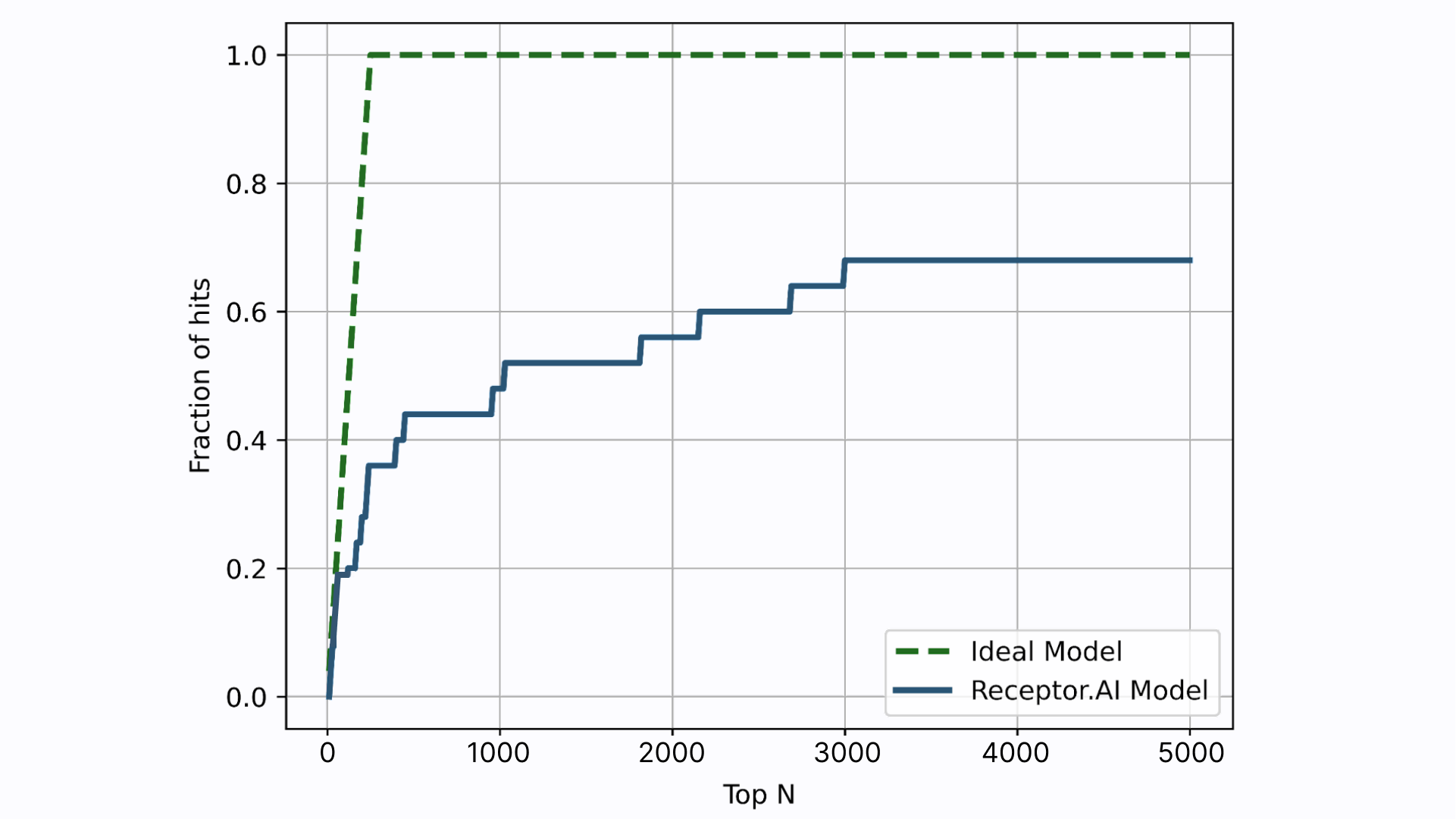 Figure 7. Enrichment plot for the known FADS1 ligands mixed into the virtual screening library
Figure 7. Enrichment plot for the known FADS1 ligands mixed into the virtual screening library
The molecules at positions 2, 3, 7, and 9 in the final ranked list are known ligands with high experimentally determined activity. The other six compounds in the top 10 demonstrate higher predicted selectivity against three explicit off-targets compared to the known ligands (Figure 8). The predicted ADME-Tox parameters and the docking poses of the top three hit candidates are shown in Figure 9.
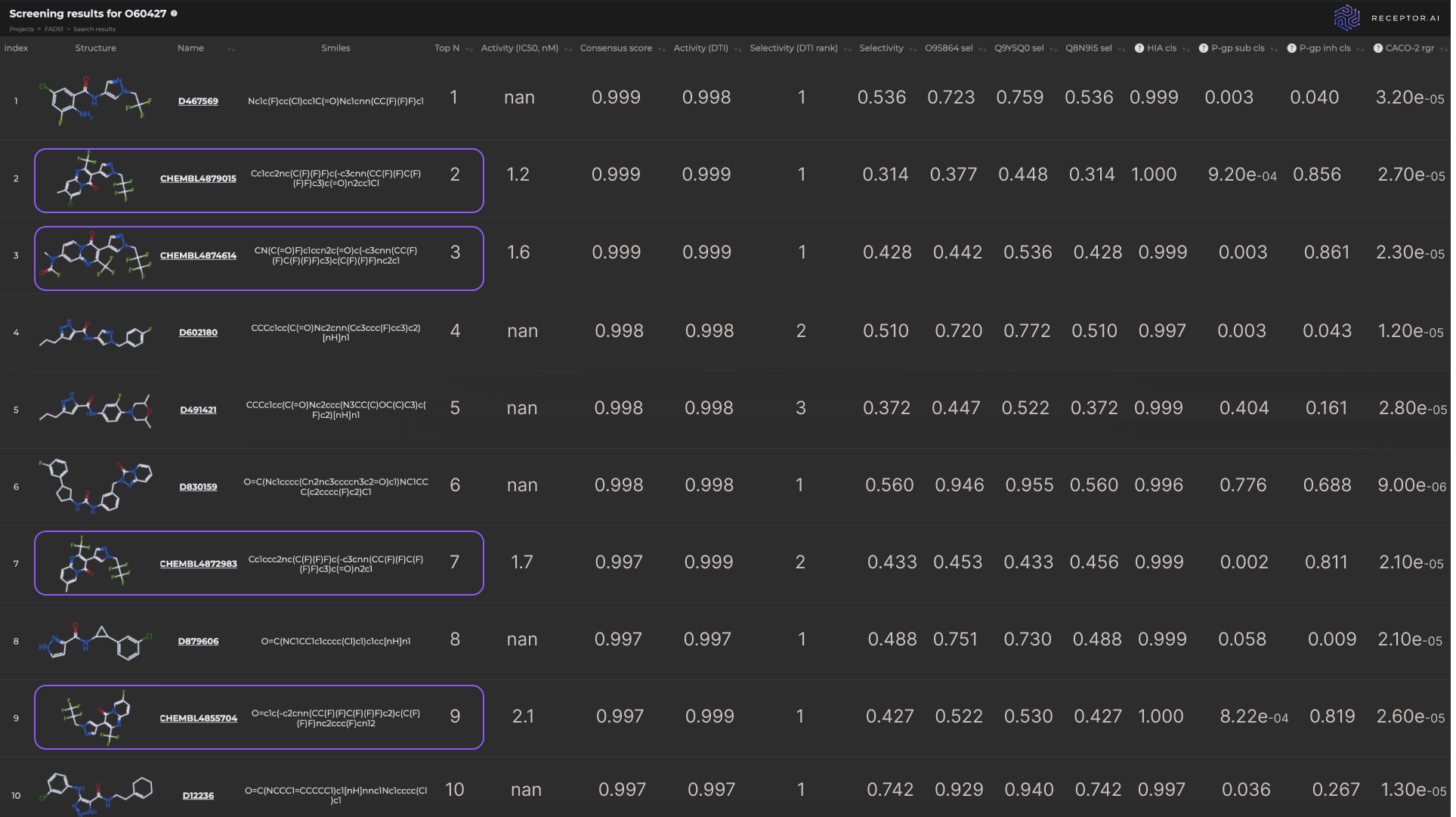 Figure 8. Properties of top 10 predicted FADS1 hit candidates
Figure 8. Properties of top 10 predicted FADS1 hit candidates
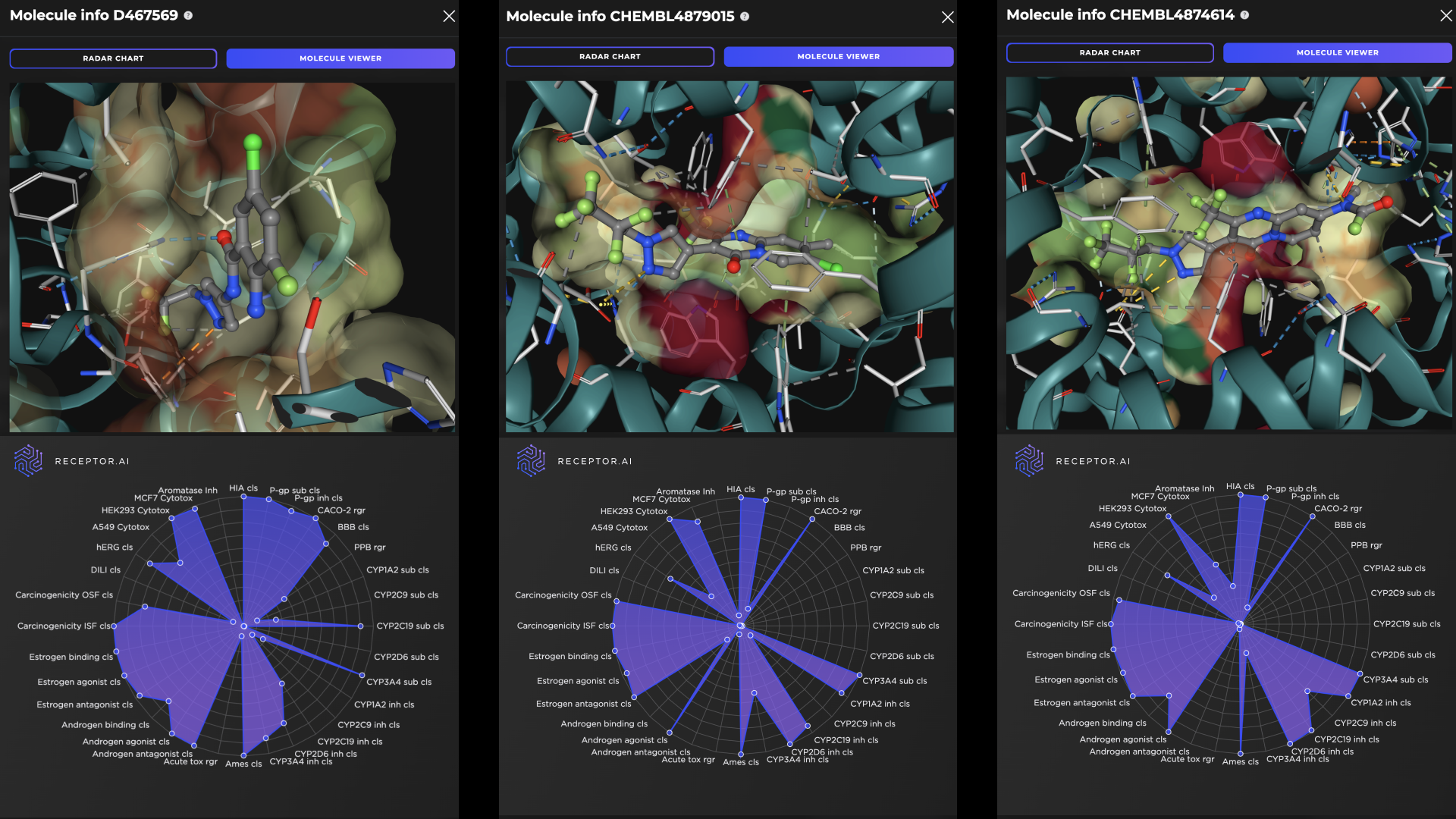 Figure 9. Docking poses (top panels) and the radar charts of ADME-Tox parameters (bottom panels) for the top three predicted FADS1 hit candidates
Figure 9. Docking poses (top panels) and the radar charts of ADME-Tox parameters (bottom panels) for the top three predicted FADS1 hit candidates
BioNeMo cloud APIs biochemistry foundation AI models could be leveraged by Receptor.AI to build applications for in silico drug discovery services. Examples include prediction of the protein structure, identification of the binding pockets, protein featurization, molecule generation, virtual compound library screening, and performing AI-based molecular docking.
The final list of 10 predicted candidates includes four experimentally validated ligands for the protein of interest, which indicates high practical relevance of the proposed pipeline.
Not only were known ligands successfully identified among the top hit candidates, but new hit candidates also emerged with potential high activity and selectivity. This indicates the utility of the proposed pipeline in discovering prospective hit compounds with novel molecular scaffolds.
Conclusion
A significant finding for Receptor.ai is the cost-saving aspect of integrating accelerated computing into the virtual screening pipeline. This change resulted in a dramatic run-time reduction and an overall 49% drop in the final cost, from $0.43 to $0.22 per instance-hour. This showcases the economic and efficiency benefits of leveraging accelerated computing in large-scale computational drug discovery campaigns, providing a more cost-effective and speedy model for virtual screening to the industry and academia.
The next steps of this exciting collaboration will involve the integration of BioNeMo Framework to train Receptor.AI models with proprietary data and Receptor.AI advanced selectivity workflow. We aim to produce the ensembles of representative protein conformations by running molecular dynamics simulations on NVIDIA high-performance, accelerated workstations and cloud instances. Further development of this technology stack will target creating high-performance AI-based drug discovery platforms for a wide range of biotech and pharma companies.
Learn more about accelerating AI-powered drug discovery using Cloud APIs to harness powerful, customizable generative AI without the complexity of infrastructure management. Get started with NVIDIA BioNeMo.
Source:: NVIDIA
