
Diamond Light Source is a world-renowned synchrotron facility in the UK that provides scientists with access to intense beams of x-rays, infrared, and other…
Diamond Light Source is a world-renowned synchrotron facility in the UK that provides scientists with access to intense beams of x-rays, infrared, and other forms of light to study materials and biological structures. The facility boasts over 30 experimental stations or beamlines, and is home to some of the most advanced and complex scientific research projects in the world.
I08-1, a soft x-ray beamline at Diamond Light Source, offers an advanced high-resolution imaging technique called ptychography to provide nanoscale-resolution images. Ptychography uses a computational imaging approach which, from measurements or diffraction patterns resulting from the interaction of an x-ray beam with a sample, can reconstruct an image of the sample to nanometer-scale resolution.
This is critical for imaging nanometer-scale features in many biological structures, such as mitochondria and organelles in cells, and also the interior structure and defects in material science samples. This process of reconstructing an image is powerful but can result in a significant gap between measuring the data and seeing an image.
The I08-1 detector operates in the range of 25 image frames per second, but detectors that can operate at the thousands of image frames per second range are on the horizon. Accelerated computing at the edge is needed for these sensor instruments.
Faster scans enable the study of more dynamic working processes. They increase the throughput of experiments, and live processing offers users real-time feedback to adjust the experiment samples, detector settings, and explore the sample to discover interesting scientific results.
This post discusses our work with I08-1 to speed up the live processing of beamline experiment data by refactoring the data analysis workflows. It also addresses key challenges, such as the generic ptychography workflow that currently operates in a serial fashion where images are written to disk at a frame rate frequency of 25 Hz.
The live-processing pipeline is launched after the scan is finished, and the processing application (PtyPy) can work on the complete dataset. The PtyPy application is optimized using GPU acceleration, but I/O communications are a major bottleneck to achieving higher throughputs.
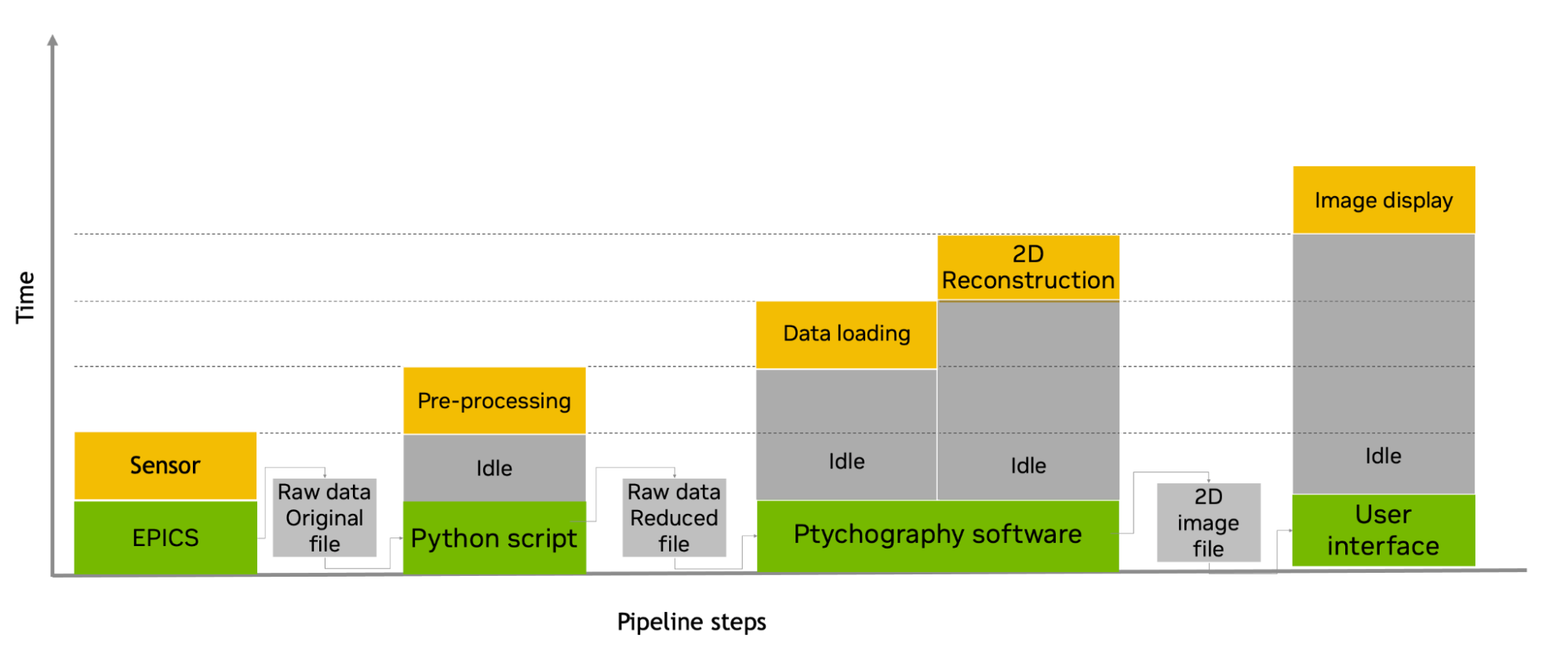 Figure 1. Serial file-based ptychographic pipeline before introduction of NVIDIA Holoscan
Figure 1. Serial file-based ptychographic pipeline before introduction of NVIDIA Holoscan
To accelerate the ptychography workflow, we incorporated NVIDIA Holoscan, an SDK for sensor processing that makes it easier for scientists, researchers, and developers to optimize and scale sensor processing workflows (Figure 2). For example, the JAX library was used within a Holoscan operator to speed up the image preprocessing.
Holoscan enables researchers and developers to develop high-performance, low-latency sensor processing applications that can scale more easily using reference examples in familiar languages.
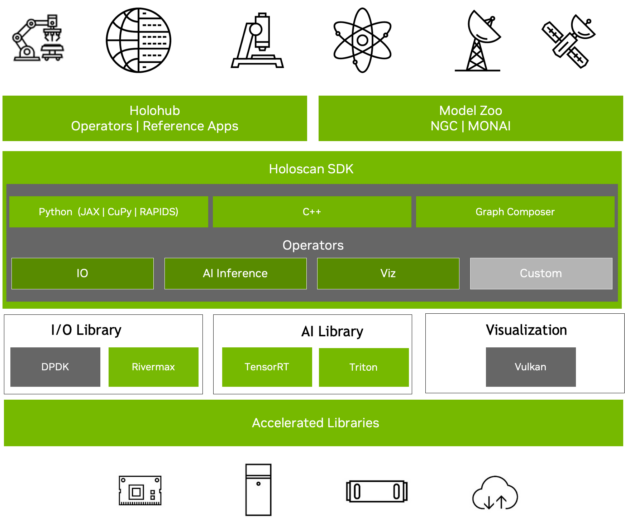 Figure 2. NVIDIA Holoscan SDK is designed for sensor applications, from surgery to satellites
Figure 2. NVIDIA Holoscan SDK is designed for sensor applications, from surgery to satellites
The soft x-ray ptychography instrument at I08-1 uses a sCMOS camera for collecting diffraction data for the ptychography experiments. The raw data comes as frames of shape (2048, 2048) and type uint16. Before feeding this data into the iterative ptychography solver application (PtyPy), the following routine preprocessing tasks are performed per raw frame:
- Subtraction of a background dark current image
- Cropping around the center and re-binning to reduce reconstruction times
While the first task is necessary to obtain clean diffraction images, the other two provide a level of loss-less compression depending on experimental circumstances. Ideally, all of these steps are performed as close as possible to the source (on-chip or using FPGAs, for example). Unfortunately, none of these are available options for the particular camera used in this scenario.
JAX was used to significantly speed up a single-threaded Python script performing the tasks listed above, and with minimal changes to the code. Because the original frame processing code was written in NumPy, the JAX JIT was able to fuse the processing routine into a single GPU kernel, which yielded a speedup of more than 2,000x for a single image over the original NumPy version (ignoring the required data transfers from host to device). Even with data transfers, the speedup is more than 40x over the original CPU-based NumPy implementation.
While acquiring the data can be relatively quick, it can easily take minutes or tens of minutes to reconstruct the data into an image that the investigator or researcher can interpret. This deadtime between a scan and an image is both inefficient and hampers the ability of the investigator to determine if the instrument has been set up properly or if the sample area being scanned is interesting until a scan result is shown.
By architecting the ptychography application as a collection of application and Holoscan operator fragments, developers can leverage the PtyPy ptychography code, Holoscan AI inference, and Holoscan network operators, to relatively quickly prototype and test a new GPU-accelerated version of the live-processing ptychography application.
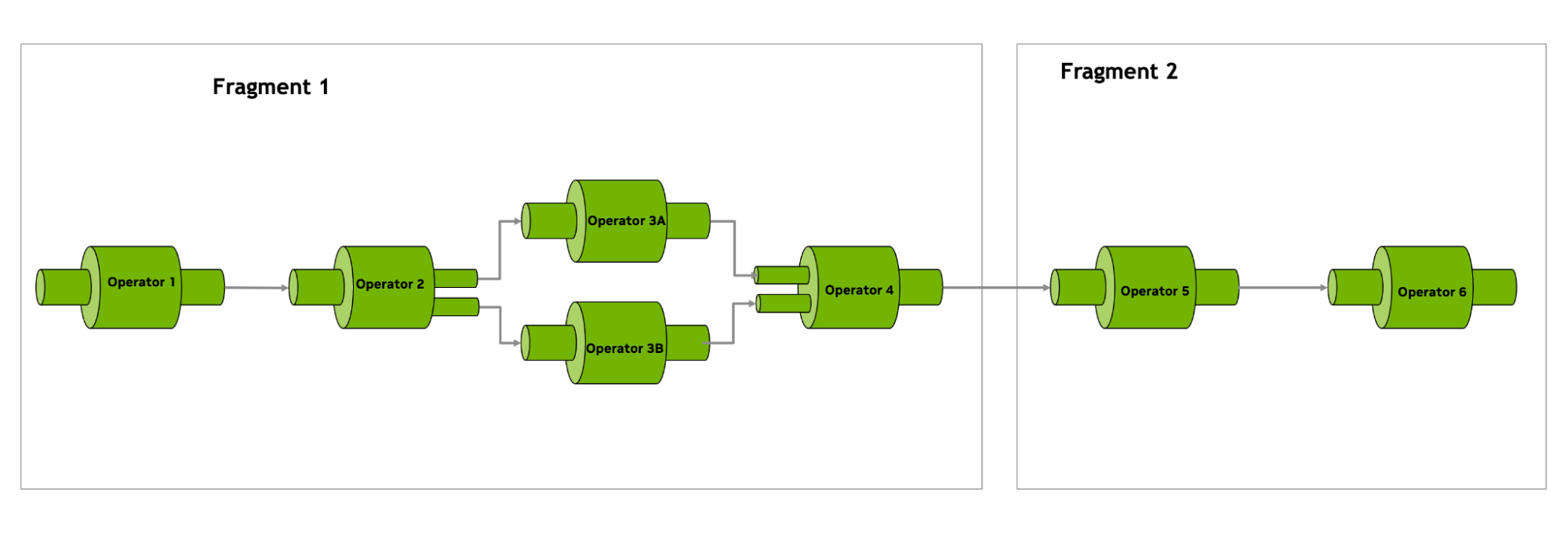 Figure 3. Applications in Holoscan are a directed acyclic graph of operators
Figure 3. Applications in Holoscan are a directed acyclic graph of operators
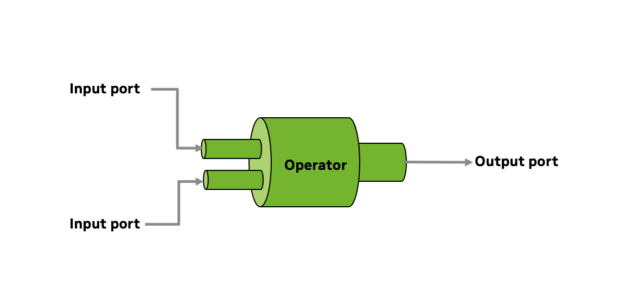 Figure 4. Operators ingest input data, then process and publish on the output ports
Figure 4. Operators ingest input data, then process and publish on the output ports
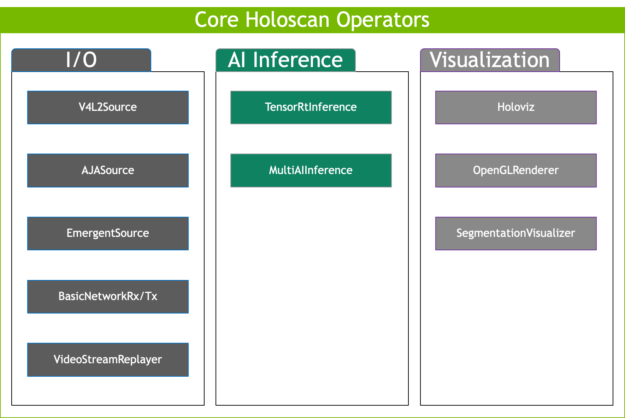 Figure 5. Holoscan SDK reference applications and core operators facilitate streaming edge HPC development
Figure 5. Holoscan SDK reference applications and core operators facilitate streaming edge HPC development
The next part of the challenge was how to speed up the live-processing frame rate for the beamline I-08 ptychography workflow to cope with the current and future sCMOS frames rates. By overlapping the serial workflow steps and using Holoscan, this beamline should be able to provide live processing that matches the frame rate of the sensor. This would enable beamline users to observe ptychographically reconstructed images of a sample in real time.
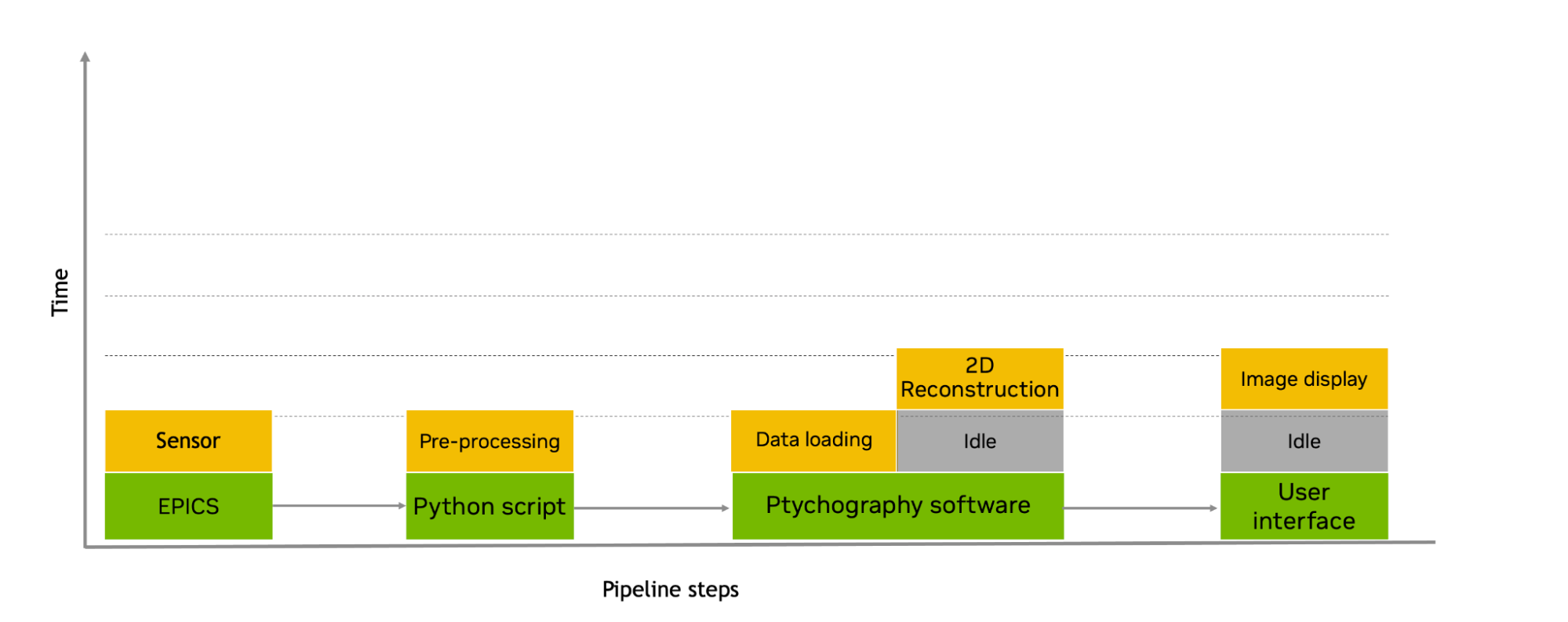 Figure 6. On-the-fly socket-based ptychographic reconstruction pipeline after using Holoscan
Figure 6. On-the-fly socket-based ptychographic reconstruction pipeline after using Holoscan
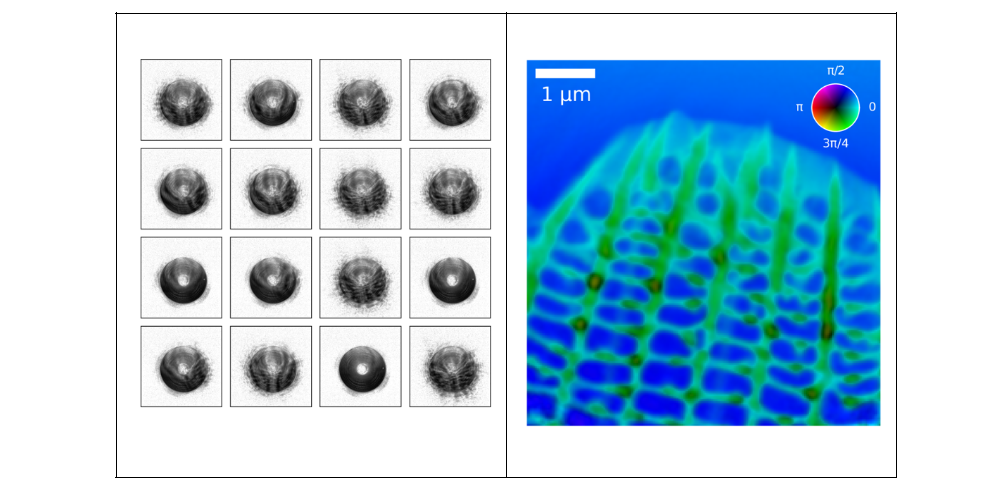 Figure 7. 2D reconstruction from 1,257 diffraction patterns to create a high-resolution image of a butterfly wing
Figure 7. 2D reconstruction from 1,257 diffraction patterns to create a high-resolution image of a butterfly wing
Before After Data collection57 sec57 sec Preprocessing94 sec58 secPtyPy data loaded119 sec61 secPtyPy data reconstructed128 sec72 secUser wait time~ 71 sec15 secTable 1. All times are relative to the start of scan except the user wait time, which is relative to the end of scan
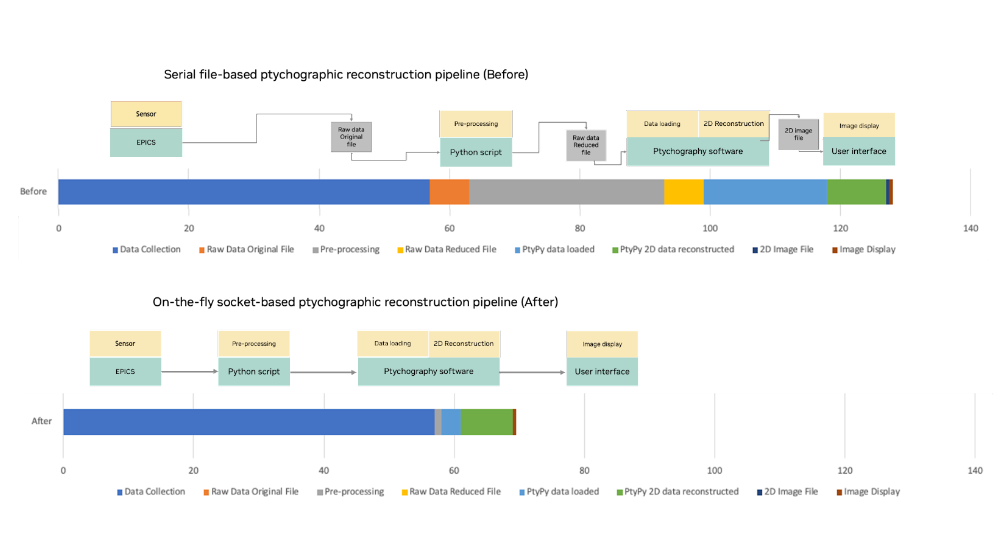 Figure 8. Comparison of file-based and streaming ptychography pipelines using Holoscan
Figure 8. Comparison of file-based and streaming ptychography pipelines using Holoscan
3D reconstruction creates a larger problem that has the same requirements for live-processing. Scaling up multi-GPU and multi-node processing to provide overlapping, parallel processing of many scans may be a means to keep up with the processing and I/O requirements for live processing.
Our collaboration aims to test various workflow configurations on a local edge server with two NVIDIA A2 GPUs, where preprocessing runs on one GPU, and image reconstruction runs on the second GPU. This approach enables focusing on bespoke ptychography code while leveraging an edge network I/O operator and AI acceleration libraries that can readily be reused and scaled up to multi-node if necessary for production usage.
Holoscan enables the creation of an end-to-end data streaming pipeline that enables live ptychographic image processing at the I08-1 beam line, which considerably enriches the overall user interaction. As mentioned, other Diamond Light Source beamlines operate in the kilohertz detection range, but none can perform live processing at that rate.
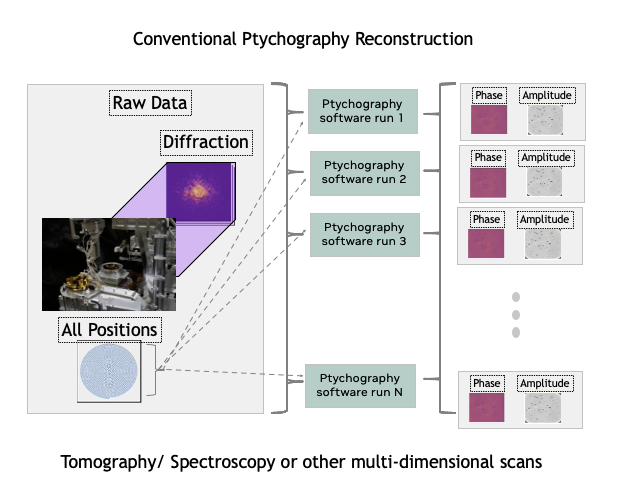 Figure 9. Tomography, spectroscopy, or other multi-dimensional scans consist of 1 to N runs of conventional ptychography software
Figure 9. Tomography, spectroscopy, or other multi-dimensional scans consist of 1 to N runs of conventional ptychography software
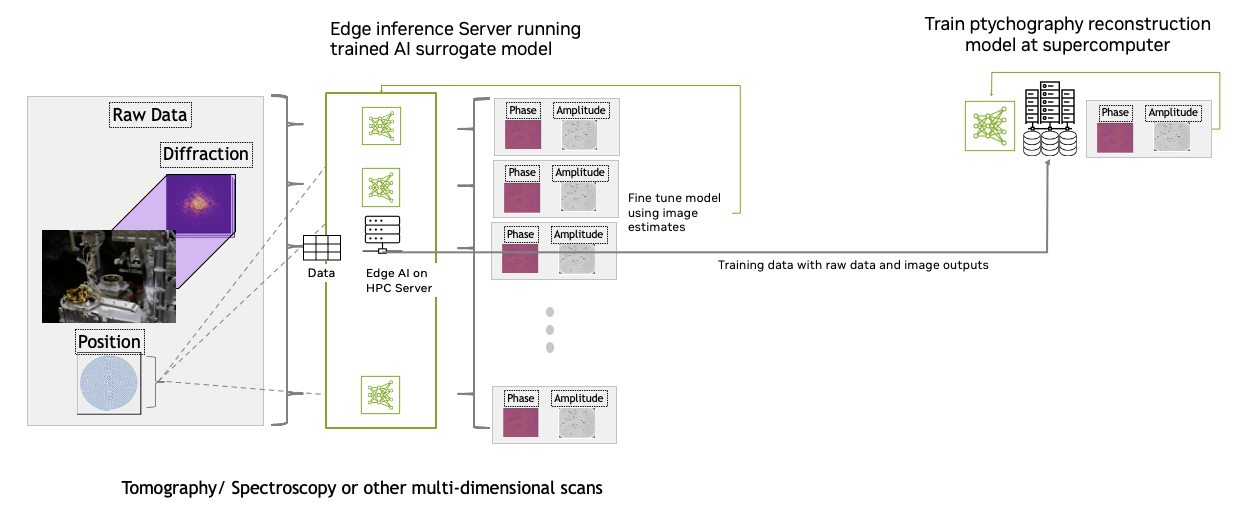 Figure 10. Train AI models for tomography, spectroscopy, or other multi-dimensional scans at the supercomputer facility
Figure 10. Train AI models for tomography, spectroscopy, or other multi-dimensional scans at the supercomputer facility
Training AI models on scan data and then using the model to run inference on GPUs at the beamline is a promising method to achieve live processing at kilohertz speeds.
Summary
Sensor processing pipelines, such as the ptychography pipeline described in this post, combine significant processing and I/O requirements into a single application. As sensor resolution and refresh rates increase, a file-based approach is no longer tenable, pushing processing redesigns to use a real-time streaming workflow.
This requires proper consideration of end-to-end performance, which immediately highlights I/O bottlenecks across the pipeline. The acceleration of the preprocessing and reconstruction operations by NVIDIA GPU edge systems using a combination of JAX, CuPy, and CUDA yields the necessary performance. However, that only amplifies the effect of the I/O bottlenecks, according to our end-to-end analysis.
Holoscan provides the tools to build streaming processing software pipelines that also leverage the capabilities of the hardware. This includes operators to ingest data directly into the GPU from the network (and the reverse) to better feed the GPUs and increase their utilization. This utilization can be particularly low during pure streaming processing like the preprocessing steps previously highlighted. Because Holoscan is designed to be easily modular, it also enables additional features like the deep learning or live-visualization features discussed.
By using GPU-accelerated computing with Holoscan, I08-1 can significantly reduce the time it takes to process x-ray microscopy data and accelerate the frame rates for image processing. The edge nodes are equipped with high-performance computing (HPC) hardware, including NVIDIA GPUs installed on CPU servers. These edge appliances or servers are designed to accelerate image processing and machine learning.
To enable real-time processing, Diamond Light Source has a distributed computing architecture that includes multiple edge nodes and a central data processing facility.
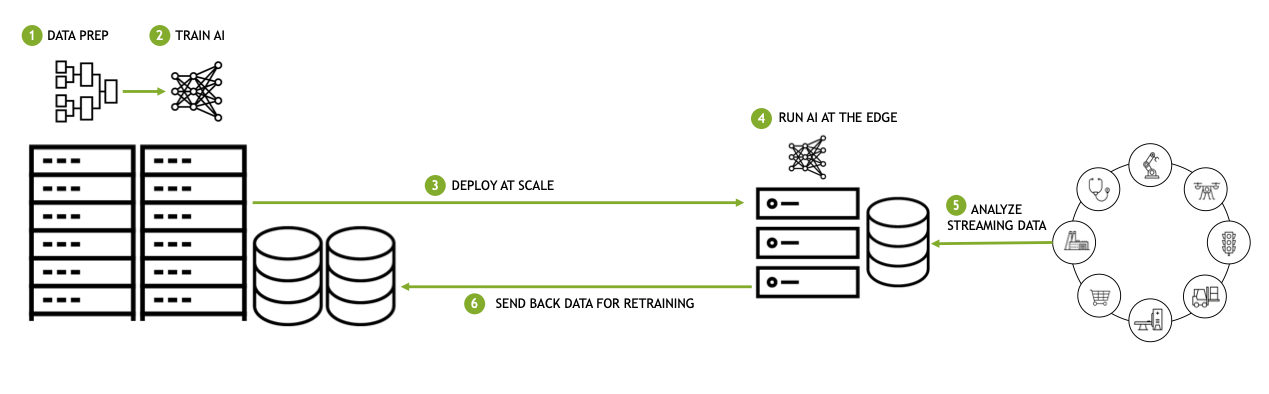 Figure 11. Data center-to-edge computing workflows run AI at the edge
Figure 11. Data center-to-edge computing workflows run AI at the edge
The edge nodes are located near the x-ray beamlines and are responsible for processing the data as it is generated. The processed data is then sent to the central data processing facility, where it’s further analyzed and stored.
Edge HPC with AI processing on NVIDIA GPUs can accelerate x-ray microscopy data processing in several ways. GPUs are highly efficient at processing large amounts of image data in parallel. In x-ray microscopy, NVIDIA GPUs can be used to accelerate tasks such as noise reduction, image registration, and image segmentation.
Machine learning algorithms, such as deep learning neural networks, can be used to analyze x-ray microscopy data and extract meaningful information. NVIDIA GPUs are well-suited for accelerating the training and inference stages of these algorithms.
By using NVIDIA Holoscan in a streaming AI framework for image processing, machine learning, tomographic reconstruction, and data compression, edge AI processing can significantly reduce the time and resources required to process x-ray microscopy data and enable faster scientific breakthroughs.
With HPC edge processing, Diamond Light Source is taking a significant step towards the democratization of science by providing scientists with the tools they need to make real-time decisions and accelerate research.
Source:: NVIDIA
