
Spark RAPIDS ML is an open-source Python package enabling NVIDIA GPU acceleration of PySpark MLlib. It offers PySpark MLlib DataFrame API compatibility and…
Spark RAPIDS ML is an open-source Python package enabling NVIDIA GPU acceleration of PySpark MLlib. It offers PySpark MLlib DataFrame API compatibility and speedups when training with the supported algorithms. See New GPU Library Lowers Compute Costs for Apache Spark ML for more details.
PySpark MLlib DataFrame API compatibility means easier incorporation into existing PySpark ML applications, with only a package import change (at most). An example of the K-means algorithm is shown below. The package import change is the only additional step necessary to enable GPU acceleration using this library.
PySpark MLlib
from pyspark.ml.clustering import KMeans
kmeans_estm = KMeans()
.setK(100)
.setFeaturesCol("features")
.setMaxIter(30)
kmeans_model = kmeans_estm.fit(pyspark_data_frame)
kmeans_model.write().save("saved-model")
transformed = kmeans_model.transform(pyspark_data_frame)
Spark RAPIDS ML
from spark_rapids_ml.clustering import KMeans
kmeans_estm = KMeans()
.setK(100)
.setFeaturesCol("features")
.setMaxIter(30)
kmeans_model = kmeans_estm.fit(pyspark_data_frame)
kmeans_model.write().save("saved-model")
transformed = kmeans_model.transform(pyspark_data_frame)
Training with supported algorithms in a benchmarking suite run in three-node Spark clusters on GPU-accelerated Databricks’ AWS-hosted Spark service demonstrated significant time and cost advantages compared to CPU-based PySpark MLlib. Specifically, this achieved a 7x to 100x speedup (depending on the algorithm) and 3x to 50x more in cost savings. Moreover, the Spark RAPIDS ML library is built on top of the proven, highly optimized RAPIDS cuML GPU-accelerated ML library.
The initial release of Spark RAPIDS ML supported GPU acceleration of a subset of PySpark MLlib algorithms with readily available counterparts in RAPIDS cuML, namely linear regression, random forest classification, random forest regression, k-means, and pca. It also included a PySpark DataFrame API for the cuML distributed implementation of exact k-nearest neighbors (k-NN) for easy incorporation of this useful algorithm into Spark applications using a familiar API.
The Spark RAPIDS ML 23.08 release includes GPU-accelerated PySpark MLlib APIs for three new algorithms:
- Binomial logistic regression with L-BFGS optimization
- Cross validation
- Uniform manifold approximation and projection (UMAP)
Binomial logistic regression with L-BFGS
Logistic regression is a well-known machine learning (ML) classification algorithm that models the conditional probability distribution of a finite valued class variable as a generalized linear function (softmax or sigmoid and linear, for example) of a feature vector.
The 23.08 release includes GPU-accelerated versions of the PySpark MLlib classification.LogisticRegression and classification.LogisticRegressionModel supporting accelerated fit and transform. This is initially for binary classification (binomial logistic regression) and L2 regularization, with full support (elastic net regularization and multi-class classification, for example) planned for upcoming releases.
Supporting accelerated logistic regression was more involved than the previously released algorithms. Unlike with the algorithms in previous releases, there was no readily available distributed implementation in cuML to leverage.
The first step was thus to contribute a multi-node multi-GPU (MNMG) extension of the cuML single GPU-accelerated L-BFGS-based logistic regression optimization algorithm (which is also used in Spark MLlib). To do this, the team followed the design pattern of the other cuML distributed implementations, with a design similar to a Message Passing Interface (MPI) centered around the GPU-optimized NVIDIA Collective Communication Library (NCCL).
The Spark RAPIDS ML bootstrapping of this implementation using the PySpark Barrier RDD and the MLlib API compatibility was then layered on top (Figure 1). As with the previously released algorithms, this design enables the GPU-accelerated distributed implementation to carry out communication in a manner that optimizes GPU utilization and over the best available interconnect between GPUs. These include Ethernet or higher-performance interconnects like NVLink and InfiniBand.
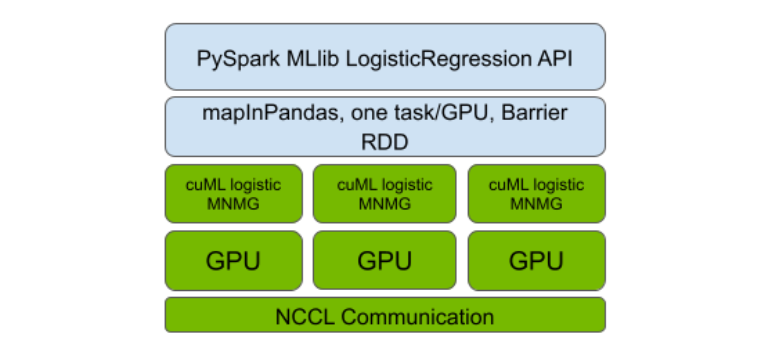 Figure 1. Integration of Spark RAPIDS ML and the newly added cuML MNMG distributed logistic regression implementation
Figure 1. Integration of Spark RAPIDS ML and the newly added cuML MNMG distributed logistic regression implementation
Performance
The benchmarking setting used for the previously released algorithms was also used to compare GPU-accelerated Spark RAPIDS ML logistic regression with the baseline CPU-based Spark ML version. The PySpark RAPIDS MLlib implementation was 6x faster and 3x more cost-efficient than the PySpark MLlib CPU implementation.
These benchmarks were run in three-node Spark clusters (one driver, two executors) on Databricks’ AWS-hosted Spark service with the hardware configurations listed below.
- In the CPU cluster, the m5.2xlarge executor and driver nodes each have eight CPU cores and 32GB of RAM.
- In the GPU cluster, the g5.2xlarge executor nodes each have the same CPU and RAM as the m5.2xlarge nodes, along with NVIDIA A10 24GB GPUs.
The benchmarks were run on a 3,000-feature 12GB synthetic dataset generated with the scikit-learn synthetic data-generating routines and stored in Parquet format on Amazon S3. Note that the runtimes are for end-to-end data loading from Amazon S3 plus fit method execution, and the spark-rapids plugin was used to accelerate data loading for GPU runs.
For more information including scripts related to this benchmark, visit NVIDIA/spark-rapids-ml on GitHub. See also a sample Jupyter Notebook demonstrating how to use the accelerated LogisticRegression APIs.
Cross validation
Cross validation is a well-known algorithm for optimizing model or training algorithm hyperparameters that are not tuned directly by the core training algorithm itself, such as the regularization parameters in logistic regression. It has long been supported in PySpark MLlib through the tuning.CrossValidator class.
Thanks to the MLlib API compatibility of Spark RAPIDS ML, the supported accelerated algorithm Estimator classes can undergo PySpark CrossValidator hyperparameter tuning out of the box. It provides speedup and cost benefits over cross validation with CPU training, which is on par with a single training run GPU compared to CPU cases. However, it suffers from the inefficiency of repeatedly copying data from CPU to GPU for each change in hyperparameter values.
Such excessive copying is a known performance bottleneck in GPU computing. It is even more pronounced for Spark RAPIDS ML, as these copies also exist between the JVM executors and Python workers over a local socket connection.
To eliminate this inefficiency, Spark RAPIDS ML now includes a specialized variant of the PySpark native CrossValidator compatible with MLlib API. It copies data to Python workers and GPUs only once while hyperparameter values change for a given cross validation fold.
The Spark RAPIDS ML specialized CrossValidator trains and evaluates all the models for the hyperparameter values under test in single respective training and evaluation Spark stages, copying the data once per stage. Figure 2 illustrates timeline traces showing patterns of copies and training and evaluation compute steps for the baseline PySpark MLlib and Spark RAPIDS ML CrossValidator versions.
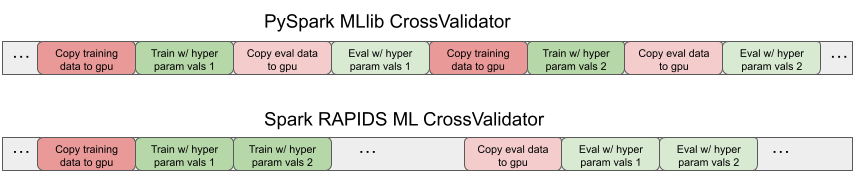 Figure 2. The Spark RAPIDS ML CrossValidator eliminates redundant data copies
Figure 2. The Spark RAPIDS ML CrossValidator eliminates redundant data copies
Performance
Our team benchmarked the new specialized CrossValidator class on three-fold cross validation with four hyperparameter values per fold for GPU-accelerated RandomForestClassifier, RandomForestRegression, and LinearRegression. We observed a 2x speedup over the baseline CrossValidator operating on the GPU-accelerated implementations.
Note that this speedup multiplies existing speedup factors due to GPU implementations of the core training algorithms when considering an overall comparison to pure CPU cross validation.
Visit NVIDIA/spark-rapids-ml on GitHub for a sample Jupyter Notebook showing the Spark MLlib API-compatible accelerated CrossValidator.
UMAP
UMAP is a state-of-the-art non-linear dimensionality reduction algorithm that is highly effective in capturing structure from the high-dimensional data into the computed lower dimensional representations or embeddings. It can be used to simplify downstream ML tasks such as classification and clustering, or for visualization.
The algorithm involves compute-intensive steps to arrive at the lower dimensional embeddings—such as k-nearest neighbors (k-NN)—in the original high-dimensional space and an iterative cross-entropy optimization of a random graph over the embeddings. It is thus a natural candidate for GPU acceleration and has been implemented in the cuML library, offering significant speedups over the original CPU implementation.
In the latest Spark RAPIDS ML release, UMAP joins exact k-NN as a non-MLlib accelerated algorithm from cuML that is wrapped in a PySpark MLlib API for easy integration in Spark applications. The design is shown in Figure 3.
The UMAP estimator’s fit method implementation is single-node and operates on a random sample of the full dataset to create a UMAP model comprising that random sample along with its embedding. The transform method of the UMAP model then extends the embedding to the rest of the dataset in a scalable, distributed manner.
It uses k-NN and cross-entropy optimization with respect to the original random sample and embedding captured in the model. The implementation overcomes serialization limitations in Spark to enable a large model size (many GBs).
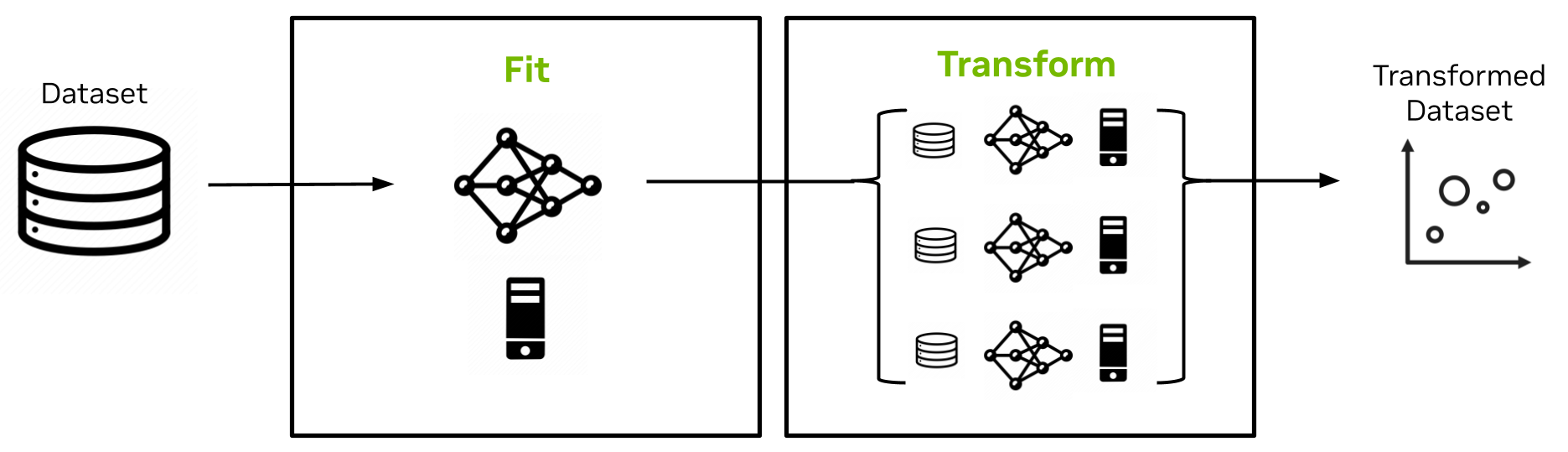 Figure 3. Spark RAPIDS ML UMAP fit and transform implementations
Figure 3. Spark RAPIDS ML UMAP fit and transform implementations
Visit NVIDIA/spark-rapids-ml on GitHub to see a sample Jupyter Notebook demonstrating GPU-accelerated UMAP on Spark. For more details about the API, see the UMAP documentation.
Summary
With Spark RAPIDS ML and its growing capabilities, you can dramatically accelerate Spark ML applications with a one-line code change while reducing your computing cost. The latest release of Spark RAPIDS ML extends these benefits of GPU acceleration to logistic regression and cross validation. In addition, GPU-accelerated UMAP is now available with a PySpark MLlib API for easier adoption in Spark ML applications.
Visit NVIDIA/spark-rapids-ml on GitHub to access Spark RAPIDS ML source code and documentation, and to provide feedback. You can also check out the resources for getting started with Spark RAPIDS ML.
Source:: NVIDIA
