
Parabricks version 4.2 has been released, furthering its mission to deliver unprecedented speed, cost-effectiveness, and accuracy in genomics sequencing…
Parabricks version 4.2 has been released, furthering its mission to deliver unprecedented speed, cost-effectiveness, and accuracy in genomics sequencing analysis. The latest version delivers a newly accelerated workflow for Oxford Nanopore sequencing (in the featured image), enables Parabricks to be run on the latest NVIDIA GPUs, and furthers Parabricks’ accelerated deep learning variant calling initiative to support data types from all major sequencer types.
Analyzing a long-read whole genome in under an hour
Parabricks v4.2 includes upgraded WDL and NextFlow workflows, as best practices for deploying Parabricks tools, available on the Parabricks Workflows GitHub repo and including both short– and long-read workflows.
This latest release of Parabricks delivers an updated Oxford Nanopore germline workflow, delivering high-speed analysis on NVIDIA H100 GPUs.
Following on from the success of the Ultrarapid Nanopore Analysis Pipeline (UNAP) released by NVIDIA in 2022, this new workflow includes the basecalling, alignment, and small and structural variant calling steps. It has updated software from Guppy to Dorado, and from PEPPER-MARGIN-DeepVariant to the newly integrated long-read variant calling of DeepVariant 1.5, deployed with Parabricks v4.2.
Figure 1 shows the workflow for the Oxford Nanopore germline sequencing analysis.
Figure 1. Workflow schematic for analysis of Oxford Nanopore germline sequencing data
This latest Oxford Nanopore workflow was recently benchmarked by Oracle Cloud on eight NVIDIA H100 GPUs, achieving an end-to-end runtime of under an hour on a single 55x coverage whole genome.
High-speed Oxford Nanopore sequencing and Parabricks workflows also have the potential to provide rapid turnaround times to clinical sequencing.
In terms of ongoing development, this workflow will be further benchmarked and optimized as part of a collaboration between NVIDIA and the Clinical Long-read Genome Initiative (lonGER) consortium. This consists of four institutes across Germany aimed at optimizing the analysis of nanopore data both in time to results and in clinical-grade accuracy of methods, to identify the most relevant clinical genomic alterations.
The National Institutes of Health Center for Alzheimer’s and Related Dementias (CARD) has developed a protocol for highly accurate, whole-genome sequencing at scale. That example study, among others, shows how Oxford Nanopore sequencing and rapid analysis can provide a comprehensive view of haplotype-resolved variation and methylation.
In a recent Nature Methods paper, the CARD team described how this makes large-scale, long, native DNA sequencing projects feasible due to the lower cost and higher throughput of Oxford Nanopore’s PromethION when compared with alternative sequencing methods.
High-accuracy variant calling for all sequencers with optimized DeepVariant models
DeepVariant, the CNN-based, high-accuracy germline variant caller, is accelerated on GPUs as part of Parabricks.
Recently, Parabricks v4.1 introduced an accelerated framework for re-training the underlying CNN model, to more easily enable custom models, and bring more accurate variant calls to analysis workflows. This brings greater accuracy by learning the error profiles of different sequencers, or the unique artifacts introduced in different high-throughput labs.
Parabricks v4.2 now comes with accelerated models pretrained for a variety of sequencer data types, as part of DeepVariant in Parabricks:
- Illumina
- Oxford Nanopore
- PacBio
- Ultima
- Singular
- …and more
The acceleration factors of these models can reach over 80x acceleration, from hours on CPU instances to under 4 minutes on NVIDIA GPUs.
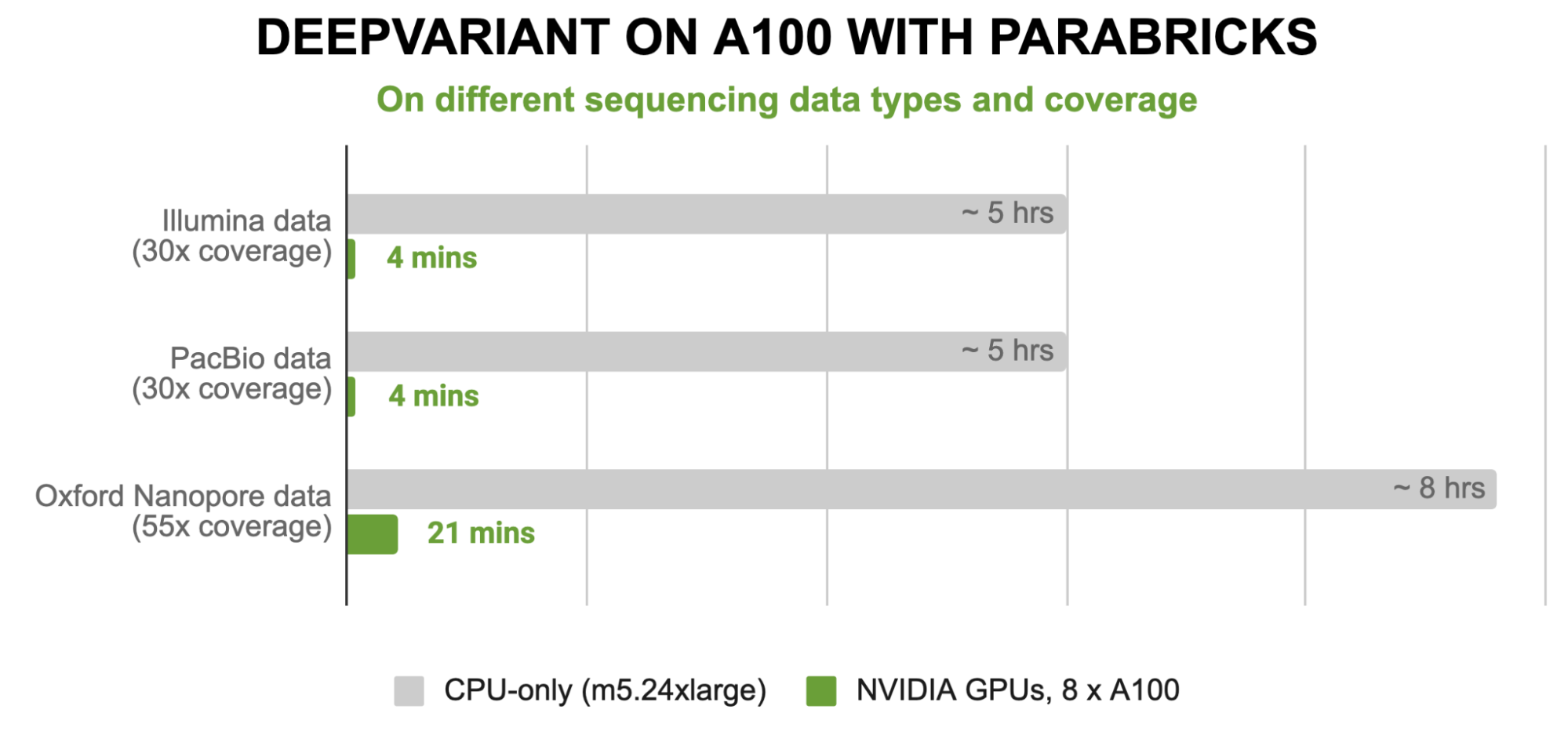 Figure 2. Runtime performance of DeepVariant in Parabricks on the NVIDIA DGX A100 compared to a cpu-only M5.24xlarge (96 vCPU cores) instance
Figure 2. Runtime performance of DeepVariant in Parabricks on the NVIDIA DGX A100 compared to a cpu-only M5.24xlarge (96 vCPU cores) instance
Unprecedented speed on NVIDIA GPUs
In high-throughput settings, moving genomic analysis workflows to GPU with Parabricks results in hugely reduced processing time.
One example of this is in Cancer Research UK’s TRACERx EVO, the latest project of TRACERx, which is the world’s largest long-term lung cancer research program, and is driven by infrastructure at the Francis Crick Institute, University College London, and the University of Manchester.
Initial results from the Francis Crick Institute show that the end-to-end analysis of whole human genomes (including FastQ alignment and deep variant calling) can be done in just over 2 hours with NVIDIA Parabricks, compared to approximately 13 hours on their NEMO CPU cluster. This performance gain is anticipated to be pushed even further on their latest GPU cluster.
For the TRACERx EVO project alone, they estimate this will save nearly 9 years of bioinformatics processing time, an improvement described as a “game-changer in terms of the feasibility of the analysis pipelines for the project,” by Mark S. Hill, principal research fellow at TRACERx EVO.
For the latest GPU architectures, the newest NVIDIA Hopper architecture has been dubbed the engine of the world’s AI infrastructure, with an order-of-magnitude performance leap for a diverse range of workloads.
High-performance computing applications being run in data centers benefit from NVIDIA Hopper’s multi-GPU scalability, and its advancements in tensor core technology, meaning impressive results such as 30x acceleration in AI inference over previous generations.
For genomics specifically, NVIDIA Hopper architectures include new dynamic programming instructions (DPX) designed to solve complex recursive problems. Dynamic programming is used commonly across multiple fields such as in graph analytics or in route optimizations. This includes in genomics with the Smith-Waterman algorithm, which underlies most aligners and multiple variant callers. The new DPX instructions accelerate these algorithms by 40x compared to CPU-only, and 7x compared to the previous NVIDIA Ampere architecture.
Combining all these advances means that the latest NVIDIA GPU architectures are incredibly well-suited to accelerate bioinformatics tools like the BWA-MEM aligner, which can run in just 8 minutes on eight NVIDIA H100 GPUs, or the deep learning–based DeepVariant variant caller, which can run in just 3 minutes on eight H100 GPUs. These runtimes mean an end-to-end germline workflow can be achieved in just 14 minutes with H100 GPUs and Parabricks.
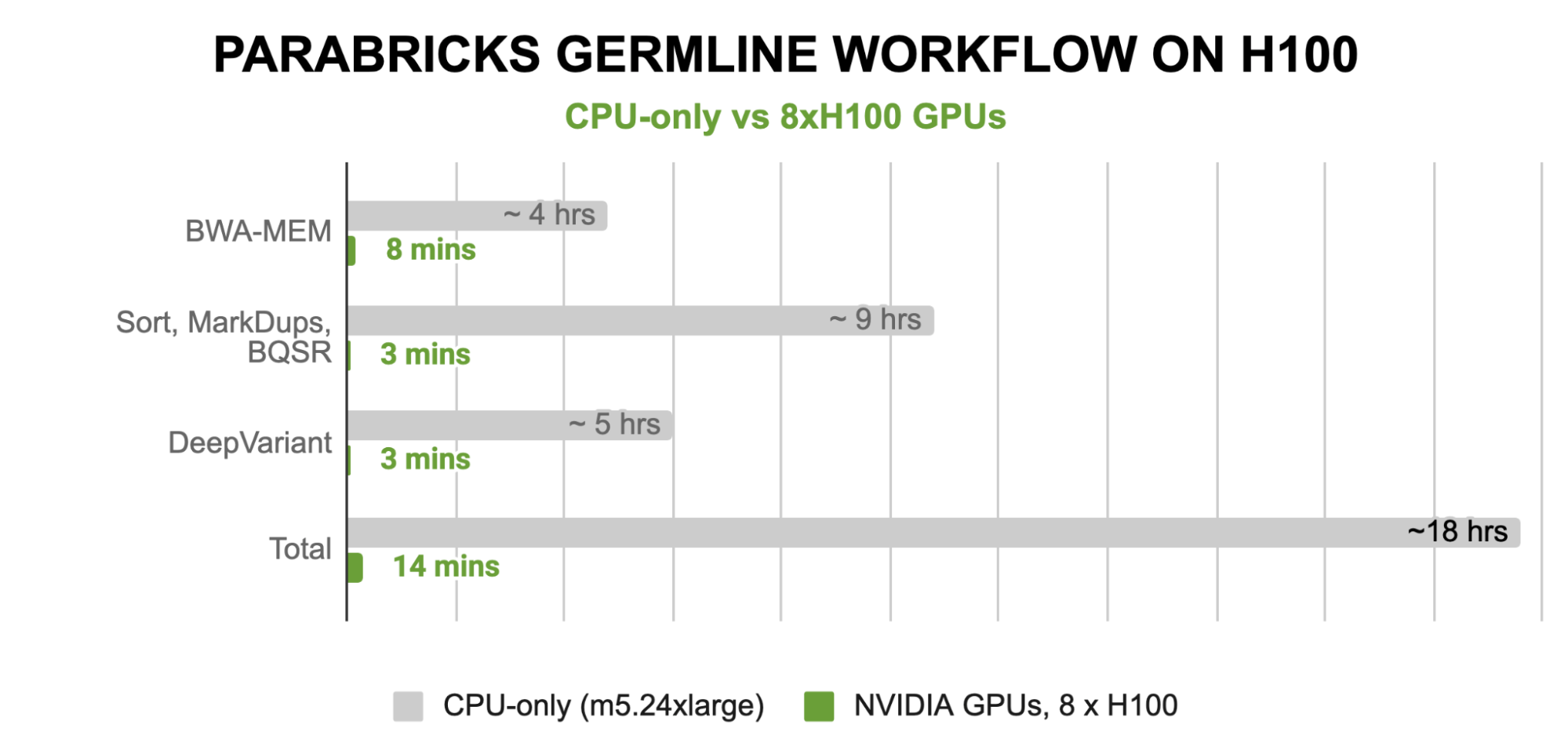 Figure 3. Runtime performance of the Parabricks germline workflow on 8 x NVIDIA H100 GPUs compared to a cpu-only M5.24xlarge (96 vCPU cores) instance
Figure 3. Runtime performance of the Parabricks germline workflow on 8 x NVIDIA H100 GPUs compared to a cpu-only M5.24xlarge (96 vCPU cores) instance
NVIDIA Parabricks v4.2 now available on NGC
Parabricks v4.2 integrates seamlessly into genomics workflows, with continued support for GPU-accelerated versions of well-established workflows with tools like BWA-MEM, and GATK and the ability to quickly train custom models for DeepVariant variant calling. In providing these for new GPU architectures, and across both short– and long-read sequencing devices, Parabricks is a truly universal full-stack acceleration platform, for gold-standard genomics analysis on GPU.
The Parabricks v4.2 container is freely available now under the NVIDIA Parabricks Collection on NGC. For WDL and NextFlow reference workflows, see the Parabricks Workflows GitHub repo.
For more information about Parabricks, see Whole Genome Sequencing Analysis, which includes customer success stories with analysis at scale, deployment in sequencers and devices, and cutting-edge research.
If you require enterprise support, contact NVIDIA sales to access enterprise benefits, including access to NVIDIA experts to ensure optimization at scale, guaranteed critical support response times, and enterprise training services
For more information about what’s new, tutorials, and deployment guides for cloud service providers, see the Parabricks documentation.
For more information about scaling sequencing analysis with Parabricks, see the NVIDIA DGX BasePOD solutions for genomic sequencing whitepaper.
Source:: NVIDIA
