King’s College London, along with partner hospitals and university collaborators, unveiled new details about one of the first projects on Cambridge-1, the UK’s most powerful supercomputer.
King’s College London, along with partner hospitals and university collaborators, unveiled new details today about one of the first projects on Cambridge-1, the United Kingdom’s most powerful supercomputer.
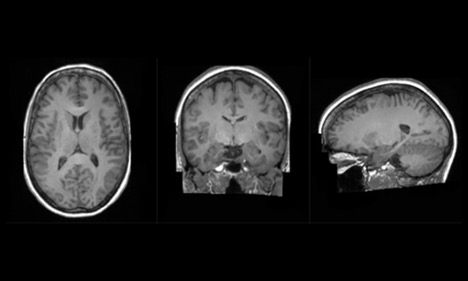
The Synthetic Brain Project is focused on building deep learning models that can synthesize artificial 3D MRI images of human brains. These models can help scientists understand what a human brain looks like across a variety of ages, genders, and diseases.
The AI models were developed by King’s College London, and NVIDIA data scientists and engineers, as part of The London Medical Imaging & AI Centre for Value Based Healthcare. The research was funded by UK Research and Innovation and a Wellcome Flagship Programme (in collaboration with University College London).
The aim of developing the AI models is to help diagnose neurological diseases based on brain MRI scans. They also could be used for predicting diseases a brain may develop over time, enabling preventative treatment.
The use of synthetic data has the additional benefit of ensuring patient privacy and gives King’s the ability to open the research to the broader UK healthcare community. Without Cambridge-1, the AI models would have taken months rather than weeks to train, and the resulting image quality would not have been as clear.
King’s and NVIDIA researchers used Cambridge-1 to scale the models to the necessary size using multiple GPUs, and then applied a process known as hyperparameter tuning, which dramatically improved the accuracy of the models.
“Cambridge-1 enables accelerated generation of synthetic data that gives researchers at King’s the ability to understand how different factors affect the brain, anatomy, and pathology,” said Jorge Cardoso, senior lecturer in Artificial Medical Intelligence at King’s College London. “We can ask our models to generate an almost infinite amount of data, with prescribed ages and diseases; with this, we can start tackling problems such as how diseases affect the brain and when abnormalities might exist.”
Introduction of the NVIDIA Cambridge-1 supercomputer poses new possibilities for groundbreaking research like the Synthetic Brain Project and could be used to accelerate research in digital biology on disease, drug design, and the human genome.
As one of the world’s top 50 fastest supercomputers, Cambridge-1 is built on 80 DGX A100 systems, integrating NVIDIA A100 GPUs, Bluefield-2 DPUs, and NVIDIA HDR InfiniBand networking.
King’s College London is leveraging NVIDIA hardware and the open-source MONAI software framework supported by PyTorch, with cuDNN and Omniverse for their Synthetic Brain Project. MONAI is a freely available, community-supported PyTorch-based framework for deep learning in healthcare imaging. The CUDA Deep Neural Network library (cuDNN) is a GPU-accelerated library for deep neural networks. Omniverse is an open platform for virtual collaboration and real-time simulation. King’s has just begun using it to visualize brains, which can help physicians better understand the morphology and pathology of brain diseases.
The increasing efficiency of deep learning architectures—together with hardware improvements—have enabled complex and high-dimensional modelling of medical volumetric data at higher resolutions. Vector-Quantized Variational Autoencoders (VQ-VAE) have been an option for an efficient generative unsupervised learning approach that can encode images to a substantially compressed representation compared to its initial size, while preserving the decoded fidelity.
King’s used a VQ-VAE inspired and 3D optimized network to efficiently encode a full-resolution brain volume, compressing the data to less than 1% of the original size while maintaining image fidelity, and outperforming the previous State-of-the-Art.
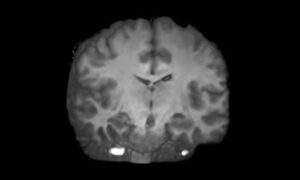 A synthetic healthy human brain generated by King’s College London and NVIDIA AI models.
A synthetic healthy human brain generated by King’s College London and NVIDIA AI models.
After the images are encoded by the VQ-VAE, the latent space is learned through a long-range transformer model optimized for the volumetric nature of the data and associated sequence length. The sequence length caused by the three-dimensional nature of the data requires unparalleled model sizes made possible by the multi-GPU and multinode scaling provided by Cambridge-1.
By sampling from these large transformer models, and conditioning on clinical variables of interest (such as age or disease), new latent space sequences can be generated, and decoded into volumetric brain images using the VQ-VAE. Transformer AI models adopt the mechanism of attention, differentially weighing the significance of each part of the input data, and used to understand these sequence lengths.
Creating generative brain images that are eerily similar to real life neurological radiology studies helps understand how the brain forms, how trauma and disease affect it, and how to help it recover. Instead of real patient data, the use of synthetic data mitigates problems with data access and patient privacy.
As part of the synthetic brain generation project from King’s College London, the code and models are open-source. NVIDIA has made open-source contributions to improve the performance of the fast-transformers project, on which The Synthetic Brain Project depends upon.
To learn more about Cambridge-1, watch the replay of the Cambridge-1 Inauguration featuring a special address from NVIDIA founder and CEO Jensen Huang, and a panel with UK healthcare experts from AstraZeneca, GSK, Guy’s and St Thomas’ NHS Foundation Trust, King’s College London and Oxford Nanopore.
Source:: NVIDIA
